We just discovered and dealt with our first major hiccups in continuing to propagate the LTEE at UT Austin. We mistakenly swapped two of the populations so that they were mislabeled. We also realized that an environmental microbe had infiltrated one population and persisted at a low enough level that our routine checks for contamination had missed this problem.
What contamination checks do we normally perform to ensure the integrity of the LTEE?
First, we look at the flasks each day to be sure that each of the cultures has only a slightly foggy whitish appearance, indicating that roughly the expected number of E. coli cells are present and that there is no obvious contamination with other microbes. Second, when we freeze down the populations every 500 generations to archive them, we plate dilutions on three different types of nutrient agar. We look for some expected patterns of colony growth on these agar plates. Cells from specific LTEE populations either grow or don’t grow on certain plates. There are also some characteristic differences in the morphologies of colonies formed by evolved cells from different LTEE populations that can sometimes be used to recognize which one is which.
These techniques can detect many problems. They have been important for maintaining the integrity of the LTEE over its long history, but they have some limitations. For example, one can only examine colonies that grow from roughly 100-300 cells in a population on an agar plate, which is a relatively small sample from populations that typically have around 300,000,000 cells after growth each day. It would also be difficult to detect contamination from microbes that don’t form colonies on any of the types of agar plates that we routinely check using these methods.
Using whole-genome DNA sequencing to check the integrity of the LTEE
Recently we started doing nanopore long-read sequencing of experiments in our lab because this lets us assemble the entire genomes of some less-studied microbes we work on (mostly insect symbionts at the moment). We’ve done a lot of short-read genome sequencing in the past, but we tend to do that in large batches to achieve economies of scale, and this means we sometimes don’t get the results for weeks or months after preparing a sample. One of the selling points of nanopore sequencing is that one can do a few samples quickly with a benchtop device. (We have been using an Oxford Nanopore MinION Mk1C.) When we froze down the 76,500-generation LTEE populations for archiving, we experimented with using nanopore sequencing as an additional way of quickly checking their integrity.
We were very glad to have given this a try because we made two important discoveries:
1. Population Switcheroo
The first thing we noticed was that two populations (A–4 and A–5) were in the wrong flasks!
These were both Ara– populations, which is one reason we didn’t detect differences in their growth on the diagnostic agar plates. The other reason is that we are still getting calibrated in terms of recognizing the colonies formed by the various populations in our lab. If we had been good at this, we probably would have noticed the mix-up sooner. We’ve started taking photos of the agar plates to better document these phenotypes. (More about that in a future post.)
To our relief, we did not detect any contamination of one population with the other in the sequencing results, meaning they were not mixed together. Instead, we must have reversed them when performing the transfers one day. Probably their positions in our tray got swapped after we removed them from the incubator. Then, we didn’t notice that the labels didn’t line up as we were pipetting.
To remedy the situation, we went back to examine earlier samples of these populations. We used PCR to screen for different ribose operon deletion alleles known to be present in all cells in each of the two evolved populations. It was clear from these result that the mix-up had happened sometime between 76,000 and 76,500 generations. We corrected the labels on our frozen samples, and these populations can now continue without interruption.
2. Acinetobacter Interloper
We also noticed a bigger problem. When we sequenced population A–3, we found that there was a sizable amount of DNA from a different bacterium. Remarkably, we got so much sequencing coverage that we were able to fully assemble the genome of the interloper and identify it as a strain of the bacterium Acinetobacter radioresistens.
It happens that we work on Acinetobacter in our lab, but only Acinetobacter baylyi. Acinetobacter radioresistens is quite different, so this wasn’t a case of contamination from a nearby lab bench. It must have randomly gotten into an LTEE flask in a different way. However, we became alarmed when we looked up more about A. radioresistens. It seems a strain of A. radioresistens was found to have survived decontamination procedures used on spacecraft that were intended to prevent us from accidentally contaminating Mars with terrestrial microbes (https://doi.org/10.1046/j.1462-2920.2003.00496.x)! It’s in the name, after all, that it is resistant, not only to radiation but to chemicals and treatments used for sterilization.
When did this contamination occur? Why wasn’t it detected sooner? We went back and looked more carefully at the “fossil record” we have frozen in the freezer and our diagnostic agar plates. We found that a few small colonies with a different appearance grew on minimal glucose plates when we spread dilutions of A–3 populations from 76,000 generations or later (see image). These slow-growing colonies were only visible when we incubated the plates for longer than normal: 48 hours versus 24 hours. Additional Nanopore sequencing of the frozen A–3 population samples found contamination in all samples with cells that formed these colonies. We did not detect any A. radioresistens DNA in the 75,500-generation sample or see any of these colonies when we plated it, so the contamination was introduced after this time point.
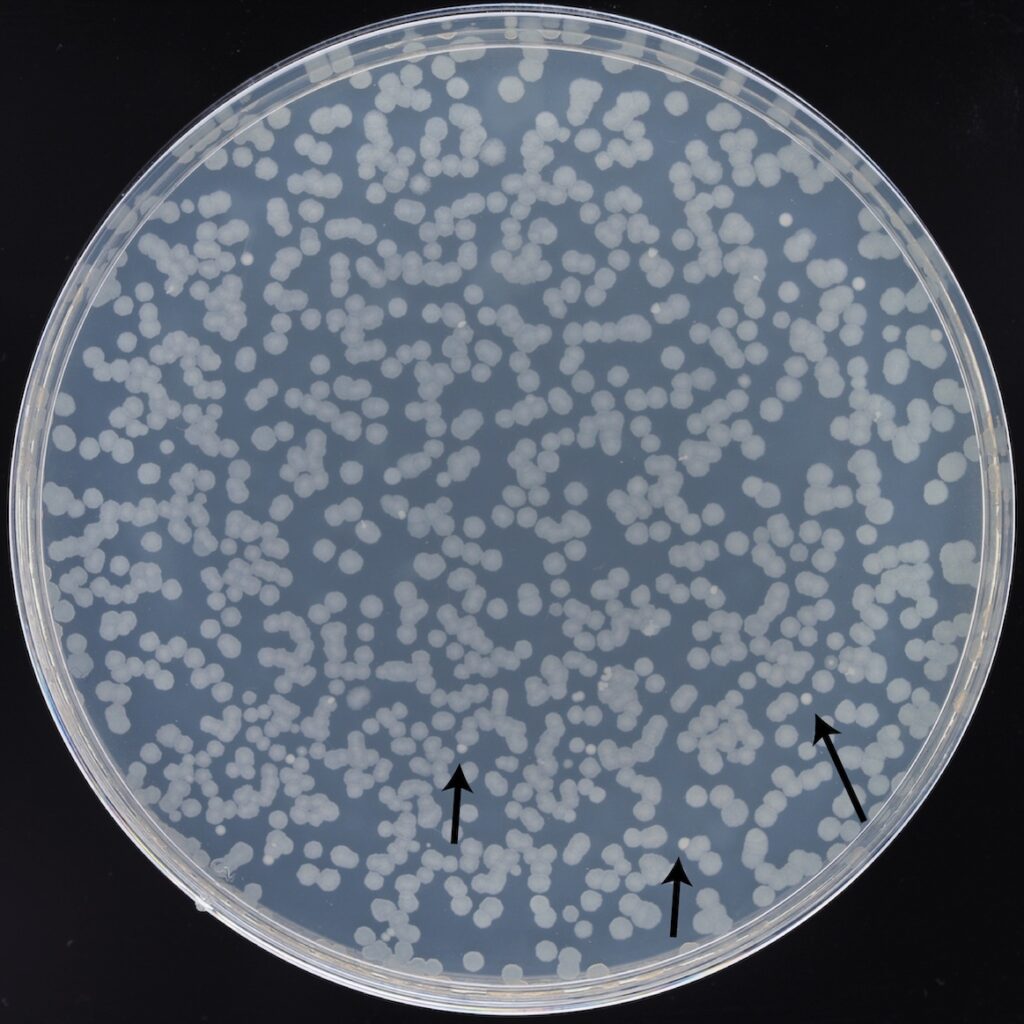
The Acinetobacter interloper
Colonies formed by Acinetobacter radioresistens cells in the 76,500-generation sample of the A–3 population can be seen on this minimal glucose agar plate after 48 hours of incubation. They are smaller and appear whiter due to being less translucent than E. coli colonies. There are ~20 A. radioresistens colonies on a plate that is crowded with >600 E. coli colonies due to the high cell density in A–3, the only population that has evolved to use citrate present in the LTEE environment. Photo credit: Jack Dwenger, Barrick Lab, UT Austin)
The solution here was to go back to the freezer and restart the A–3 population from the last “clean” sample we had. We let all of the LTEE populations continue to 77,000 generations, the next time they were set to be archived after we sorted all of this out (which took a while). Leading up to this day, we revived the 75,500 A–3 population from the freezer. Then, we substituted it in for the contaminated A–3 population so it could be in sync with their schedule. The A–3 population is now 1,500 generations behind the other 11 populations, but this isn’t the first time an LTEE population has needed to be rolled back to maintain the integrity of the experiment.
Even though this episode is now over, there are some lingering questions. Do we need to worry about A. radioresistens recontaminating the LTEE or spreading to other populations? Is it endemic in our lab? Why did it apparently invade only A–3 and remain at a low but steady frequency in this population for such a long period of time (at least 1,000 generations). We have no plans to track the answers down – it’s generally a very bad idea to purposefully grow microbes that are good at contaminating your experiments in your lab! — but we can speculate.
We noticed that A. radioresistens grows relatively slowly on minimal glucose agar that mimics the glucose-limited LTEE environment. It grows much faster on rich medium (LB). It may be that it doesn’t stand a chance in competition with most of the glucose-specialist LTEE populations and couldn’t contaminate them even if we spiked it into them. A–3 is a special case because E. coli in this population evolved to use citrate. In the process of metabolizing this abundant nutrient, they secrete various other compounds as byproducts. It’s possible the A. radioresistens persisted (at a low level) by taking over a niche that is peculiar to this population, in which it exploits one or more of these secondary nutrients better than E. coli.
Most likely this episode will remain a mystery and footnote to the LTEE—there are so many other interesting stories that have come out of these flasks in the past and that we and others are working on right now. However, we did preserve the frozen fossil record of the alternative future in which A. radioresistens co-exists with the LTEE E. coli, from sometime between 75,500 and 76,000 generations through 77,000 generations, in case anyone ever wants to study it.
Ever Onwards
The LTEE continues! Soon, we’ll be archiving the 77,500-generation samples (except A–3 will be at 76,000 generations). We will be looking with a more practiced eye at the colonies on our plates to check for possible contamination. We’ll also sequence the LTEE populations again to double-check their integrity and find less obvious problems. It will be nice to have this extra piece of mind each time that we deposit new “strata” of the “fossil record” in the freezer.
Finally, many thanks to Dan Deatherage for handling DNA sequencing and Jack Dwenger for heading up maintenance of the LTEE and taking plate photos.
Pingback: 80,000 Generations: New Olympic Record – The Long-Term Evolution Experiment
Comments are closed.