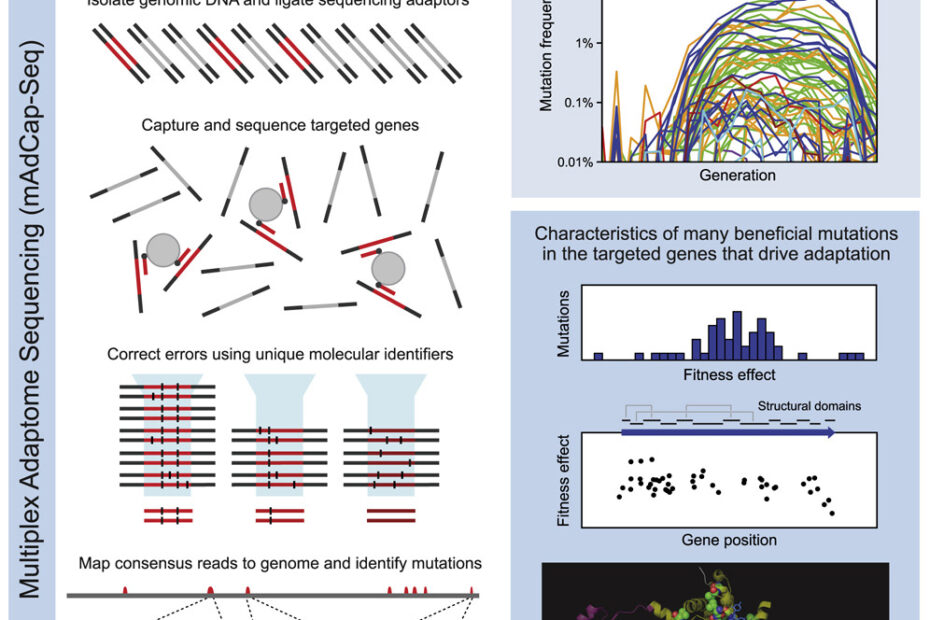
In a new paper just published in Cell Systems, Daniel Deathrage and Jeff Barrick detail an exciting approach to inferring adaptive dynamics at resolutions previously inaccessible using traditional sequencing methods.
In the past, metagenomic sequencing of microbial evolution experiments has revealed a wealth of information on the mutational dynamics of long-term evolution. For instance, in the LTEE, even if fitness trajectories decelerate over extended periods of time, mutations continue to fix in a near-constant rate. On the other hand, the time to fixation and the number of hitchhikers in the selective sweeps both increase as the population becomes better adapted.
While these insights have helped open up new avenues of interrogation, they are still limited to a regime where natural selection is dominant. This is because most whole-genome sequencing studies are limited to detecting mutations at frequencies higher than 1-10%. At this ratio, given the large population size of the LTEE, the mutation has already escaped the drift barrier and is fully under the influence of selection. Sequencing costs make it prohibitive to delve into the dynamics of mutations much below 1%.
Deathrage and Barrick circumvent this problem using an innovative solution: multiplex adaptome capture sequencing (mAdCap-seq). All 12 LTEE populations show high parallelism in the mutations they fix early in evolution; a handful genes are highly targeted. Deathrage and Barrick call these portions of genome that are particularly important to adapting to a certain environment as the organism’s ‘adaptome’. By hybridizing probes to select genes in the adaptome and sequencing those genes over a dense timecourse at very high depth, Deathrage and Barrick inferred and visualized mutational dynamics in the initial phase of the LTEE at frequencies as low as 0.01%. This allows them to study the full spectrum of beneficial mutations that arise in important genes, but are ultimately lost in the zero sum game of clonal evolution (where sex cannot recombine multiple beneficial mutations on to the same background). Indeed, in the first 500 generations, they see beneficial mutations arise that didn’t fix in the LTEE until hundreds or even thousands of generations had passed. Moreover, in 3 of the most important adaptome genes — nadR, pykF, and topA — they see unique signatures of selection on protein structure, helping understand better and perhaps even predict the spectrum of mutations that dominate early during evolution.