About a year and a half ago, I wrote about a pair of papers posted on the bioRxiv preprint website. Both papers examined the evolution of the distribution of fitness effects (DFE) in the LTEE populations, but they addressed complementary questions. One paper focused on the overall shape of the DFE as well as changes in genomic robustness to deleterious mutations and in the identity of essential genes. The other paper focused on the small, but critical, tail of beneficial mutations, and how the genes with potential beneficial mutations changed over time.
I’m happy now to announce what has become of these papers. After positive reviews and revisions, the editor at Science asked the two teams to combine their papers into a single long-format Research Article. Both teams agreed to do so, but it was a lot of work, requiring further reviews and revisions. At last, the combined paper was accepted, and it appears in the latest issue of Science. Hooray!
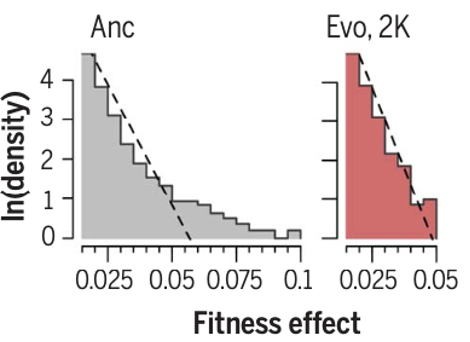
I think it’s fair to say that the paper provides an unprecedented description and analysis of how the fitness effects of mutations change over time, even under the constant conditions of the LTEE, and even as the overall shape of the DFE was little changed. As a matter of practicality for the high-throughput analyses, the paper relied on generating libraries of insertion mutations. That approach means that the new study focuses on loss-of-function mutations, which are undoubtedly important in the LTEE, but which means that mutations that refine existing functions or even generate new functions are substantially under-represented. Perhaps some future methodology will allow an even more comprehensive analysis, but the new paper provides a great start.
I want to provide a little background, not to the science, but to the interaction among the wonderful people who did the lab work—Alejandro (Alex) Couce and Anurag Limdi along with Melanie Magnan, Siân Owen, and Cristina Herren—and who led the two teams—Michael Baym and Olivier Tenaillon. When I first realized that two teams were preparing papers about the evolution of the DFEs in the LTEE, I had an immediate pang of guilt. When people ask me for LTEE strains, I ask them why they want the samples. That way, I can let them know about potential work that others are doing, which might cause someone’s work to get ‘scooped’ by another team. However, I don’t always remember all the projects, or I might fail to see the connection in the words people use to describe their plans, and people’s projects may also change after they start their work. In any case, thanks to Tanush Jagdish for letting us know that the two teams were pursuing related projects.
After that, the two teams soon began having zoom meetings to openly share and explain their questions, hypotheses, methods, and findings. There was some overlap, of course, but I think that’s valuable in science, because it indicates broad interest in the topic and, moreover, shows that the results are robust. Thus, the decision was made to submit two separate papers as joint submissions to the same journal. In a thread on social media, Michael nicely described the situation and the resulting comradery (my emphasis added):
As we were working on this we discovered that another group led by Alex Couce and Olivier Tenaillon were working on something very similar. Instead of competing we decided to talk to one another. While either group could have scooped one another, what we did instead was have some of the most fun scientific calls I’ve ever been part of, in which we discussed our work and our results, and even cross-reviewed each other’s manuscripts. We found that while we were superficially overlapping (TnSeq on the LTEE) the experiments and the questions we asked were complementary. While we’d focused on detrimental mutations after long periods, they looked at early beneficial mutations. Yet we’d found similar trends! In the end, Alex and Olivier’s insights and perspective gave us both a much deeper understanding of the science, and a lot more confidence in the results. As our papers are complementary and speak to one another, we coordinated preprinting and will be co-submitting.
More recently, as we combined the two papers into one, we wanted to leave a ‘footprint’ that would reveal the origins of the integrated study. At the start of the Discussion, we wrote:
This paper began as two separate projects performed by two different teams, using similar but not identical methods. As we discussed our findings together, we discovered that each project reinforced and complemented the other. They reinforce one another by finding the same evolution of the overall form of the DFE; they are complementary because one project delved deeply into the fine-scale genetic changes in the deleterious tail while the other did so for the beneficial tail.
We wondered if the editor might reduce or eliminate this passage, since a paper’s history doesn’t bear directly on its results. We were delighted, therefore, when we saw that the passage remained, because it sheds light on how civility, cooperation, and collaboration can succeed even in the highly competitive world of science.
Changing fitness effects of mutations through long-term bacterial evolution Journal Article
Science, 383 (6681), pp. eadd1417, 2024.
Pingback: Friday links: preregistration prevalence, superforecasters, and more | Dynamic Ecology
Comments are closed.