I’m excited to announce two new papers posted on the bioRxiv preprint website. Both studies examine the evolution of the distribution of fitness effects (DFE) in the LTEE lineages, but they address complementary questions.
The work was performed by two outstanding teams:
- Anurag Limdi, Siân Owen, Cristina Herren, and Michael Baym
- Alejandro Couce, Melanie Magnan, and Olivier Tenaillon
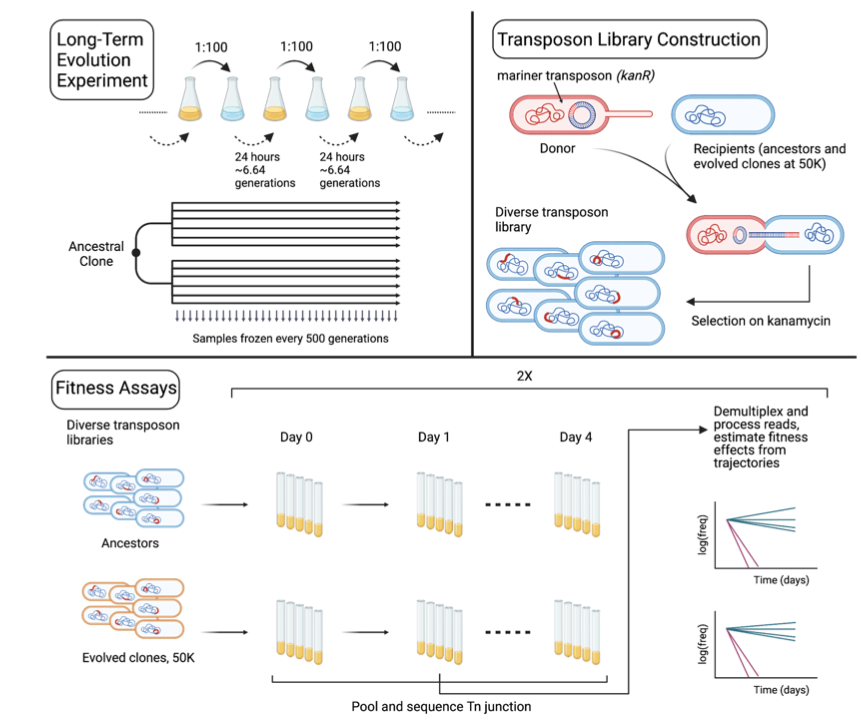
Both studies generated high-coverage transposon-insertion libraries in the LTEE ancestor and various evolved isolates. Importantly, the approach allows the identification of the insertion sites of each mutant. They then propagated the mutant libraries in the LTEE environment and tracked the frequencies of all the mutants over time—in essence, bulk fitness assays involving hundreds of thousands of mutations. The figure below, from the paper by Limdi et al., outlines the approach.
Limdi et al. first show that the overall structure of the DFE has hardly changed after 50,000 generations, contrary to some predictions from evolutionary theory.
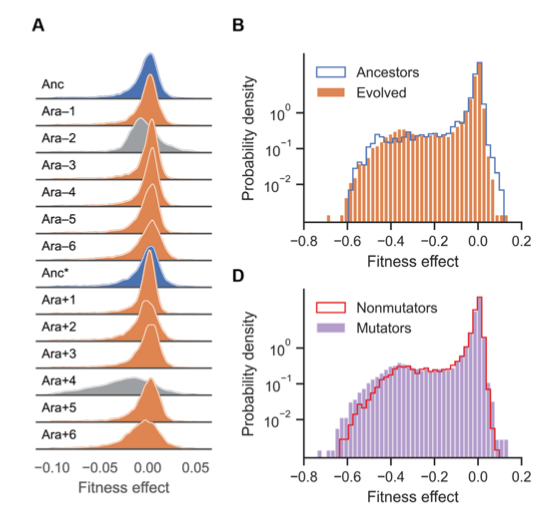
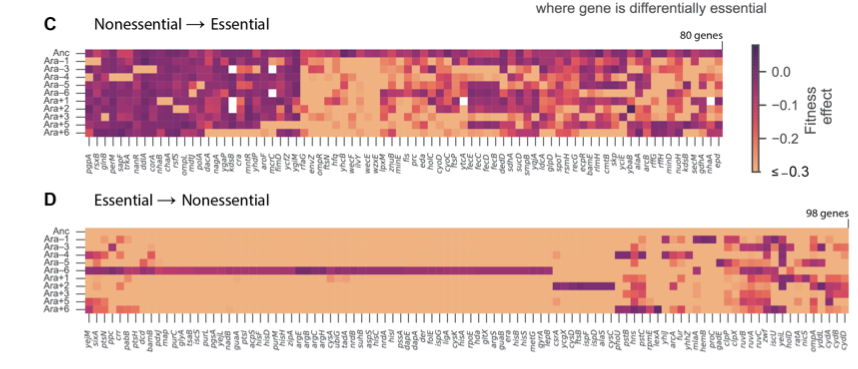
Limdi et al. also discover that the identity of essential genes has changed substantially, and often in parallel across multiple LTEE lineages.
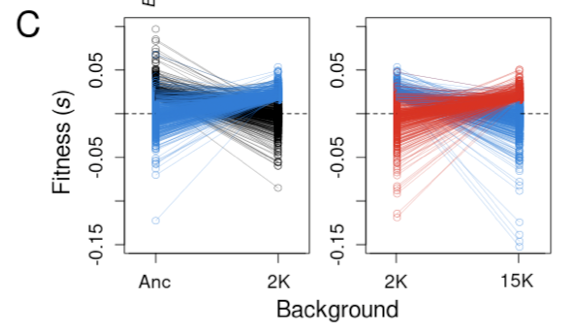
Couce et al., by contrast, focus their attention on the small tail of the beneficial mutations that drive adaptive evolution. They show that the beneficial tail becomes much smaller over time and approaches an exponential distribution.
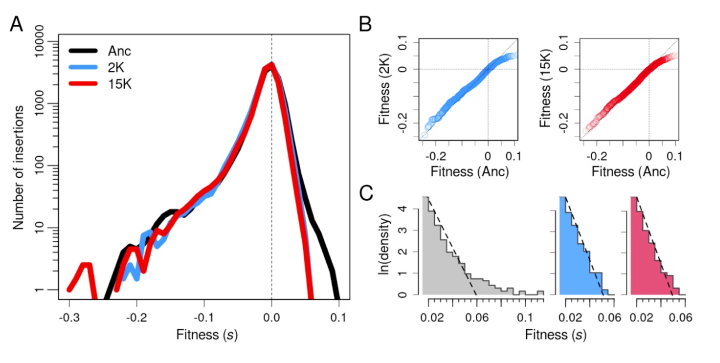
Couce et al. also demonstrate rapid turnover in the identity of the genes that harbor potential beneficial mutations.
Together these papers provide an unprecedented description of how the fitness effects of the same mutations can change over time, even under the constant conditions of the LTEE. These changes radically alter the fate of specific mutations, even as the overall genomic and fitness dynamics of the evolving populations follow more predictable trajectories.
Here are links to the two preprints: