This page is part of a series of posts about what we’ve learned from the LTEE. You can view the original version on Telliamed Revisited.
Repeatability of Evolution
The LTEE has produced many striking examples of both parallel (repeatable) and divergent evolution across the 12 replicate populations, including at both the phenotypic and genetic levels.
These examples all bear on the reproducibility of evolution, which is one of the core questions that the LTEE seeks to address. The answer is not a simple one with a dichotomous “yes/no” outcome, because evolution is an intriguing mix of random (mutation and drift) and directional (natural selection) processes. But the LTEE offers the opportunity to examine this question more thoroughly than almost any other biological system studied to date.
Examples of Parallel Evolution in the LTEE
Fitness. The trajectories for fitness, as measured in the environment of the LTEE, have been very similar across the replicate populations, although they are certainly not identical (Lenski and Travisano, 1994; Travisano et al., 1995). But perhaps that’s not too surprising because fitness integrates, rather than atomizes, the underlying changes.
Cell size. One of the most strikingly parallel trends has been in the size of the individual cells. All 12 populations produce cells that are much larger than the ancestor (Lenski and Travisano, 1994). If you had asked me, I would have thought the cells should become smaller based on surface-to-volume ratio considerations in a resource-limited environment. But the bacteria obviously had a different “opinion” about this, so to say.
Genetics. And it’s not just phenotypic traits that show parallel evolution. We’ve found three genes that have fixed mutations in all 12 populations (V. Cooper et al., 2001; Woods et al., 2005), although the exact mutations at the sequence level differ in almost every case. By contrast, most of the 4,000+ genes retain the ancestral sequence in most or all of the lines because, while the LTEE is a long experiment, it’s still just a “drop in the bucket” of evolutionary time.
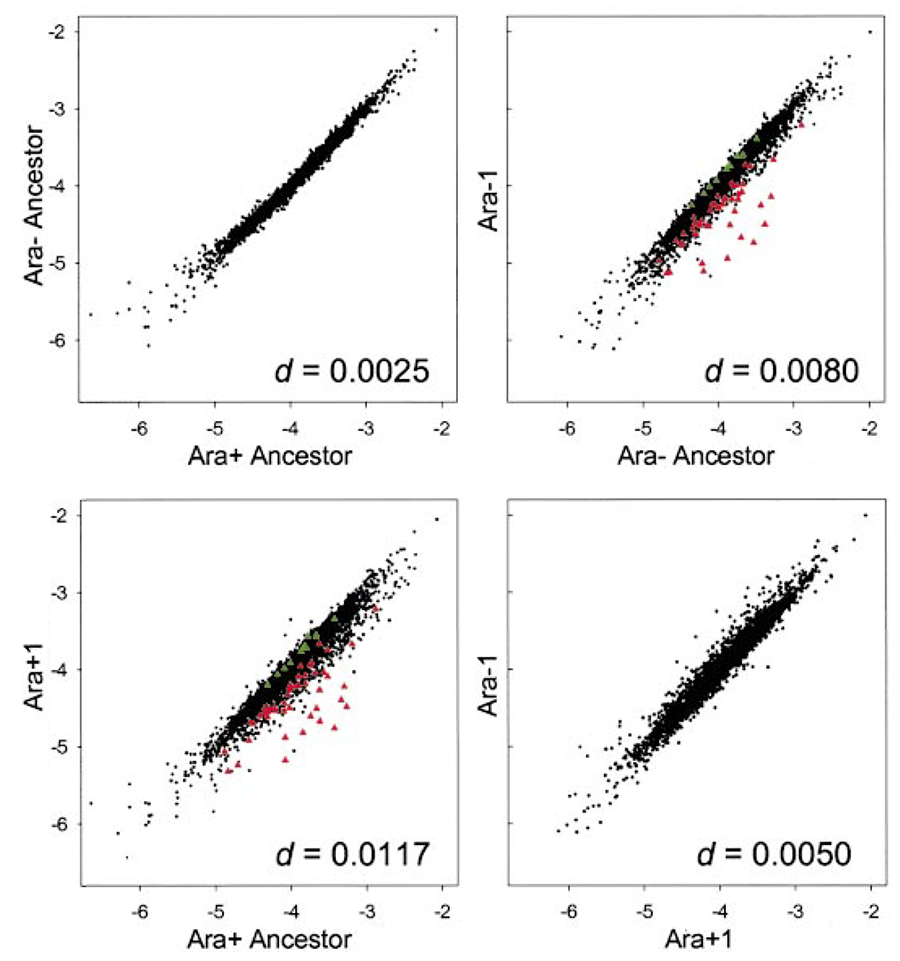
Gene expression profiles. Perhaps my favorite example of parallel evolution is at the level of changes in gene expression across the entire “transcriptome” (T. Cooper et al., 2003). We examined only two of the LTEE lines (because of costs) and we used the old approach of microarrays (as opposed to new RNAseq methods). The changes in the global expression profiles were strikingly parallel, so that after 20,000 generations (when this analysis was done) these two independently evolved lines were more alike than either was to its ancestor (see figure below). The identity of the genes whose expression changed in parallel suggested a shared underlying cause—a change in a “global” regulon, a high-level pathway that coordinately regulates the expression of many genes. From there, we tracked down a mutation in a gene called spoT, a key gene in that regulon.
The figure below shows the comparisons in global gene-expression profiles between: (top left) the ancestor and itself, as a control; (top right) one evolved line and the ancestor; (bottom left) another evolved line and the ancestor; and (top right) the two independently evolved lines relative to one another. The figure is modified from T. Cooper et al., 2003, Proc. Natl. Acad. Sci. USA; it is shown here under the doctrine of fair use.
And back to genetics. When the evolved version (allele) of spoT was moved to the ancestral genome, it conferred a significant competitive advantage, demonstrating that it was indeed a beneficial mutation. Moreover, the ancestor with the evolved spoT allele recapitulated many of the changes in gene expression that we saw in the evolved lines and that led to its discovery, which provided satisfying closure to our inferences. And when we sequenced spoT in all 12 of the LTEE lines, we found that 8 of them had substitutions in that gene. Nonetheless, a mystery remained: one of the two populations with the expression profile that evolved in parallel, and which led to the discovery of the many parallel mutations in spoT, did not itself have a mutation in spoT. A mutation in some other gene (not one of the other candidate genes we had sequenced) must “mimic” the effects of the evolved spoT mutation in the other line whose gene-expression profile we had studied. The LTEE is not only a valuable resource for studying evolution, it also generates many mutations worthy of study from molecular, genetic, biochemical, physiological, and other perspectives.
Examples of Divergent Outcomes in the LTEE
Citrate utilization. The most striking case of divergence we’ve seen is that one of the populations evolved the ability to consume the citrate that has been present throughout the LTEE (Blount et al., 2008; Blount et al., 2012). It took more than 30,000 generations for this innovation to arise in that population, and none of the other populations have figured it out even after almost 60,000 generations.
Growth on maltose and resistance to phage Lambda. There are many other, more subtle examples of phenotypic divergence. One that I find very interesting concerns the differences in adaptation to glucose and maltose (Travisano et al., 1995). Maltose is simply a dimer of glucose. Glucose is the limiting resource in the LTEE (leaving aside the one line that evolved the ability to use citrate). One might expect, therefore, that the bulk of fitness gains measured in the LTEE environment would carry over if maltose were substituted for glucose in the medium. In fact, however, that is not the case. After 2,000 generations, the variation among the replicate lines in their performance on maltose was at least an order of magnitude greater than their variation in glucose. Now some of the lines cannot grow on maltose at all. And the same mutations responsible for that complete loss of growth on maltose caused those lines to become resistant to infection by a virus, phage Lambda, even though the LTEE lines were never exposed to the virus (Meyer et al., 2010).
Inferences on Parallel and Divergent Evolution in Nature and in the Laboratory
Challenges in interpreting nature. Parallel and divergent outcomes are, of course, also seen in nature, but it is often difficult to interpret these cases. If two or more lineages underwent parallel phenotypic changes, was it because they shared genetic variation that was present before the lineages split or by later gene flow? If so, the parallel changes may not be truly independent evolutionary outcomes. And even if shared variation can be excluded (e.g., the parallel phenotypic changes have different genetic bases), what’s the relevant denominator? That is, how often did parallel evolution occur relative to how often it could have occurred? Also, if two or more lineages diverged phenotypically, does that reflect the random effects of mutation and drift? Or might it reflect instead subtle differences in the environment (ones that may be imperceptible to us, but important to the organisms) or the ancestral genotypes (i.e., divergence that occurred prior to the lineages encountering similar environments)?
Easier inferences in the LTEE. By contrast, the 12 populations in the LTEE all started from the same ancestral strain of E. coli. Although they share the same ancestor, the populations do not share genetic variation; in fact, there was no variation at the outset because each population was started from a single haploid cell. In other words, all of the variation that underlies changes we observe in the LTEE arose by new mutations that occurred during the experiment itself. And of course, the 12 populations have evolved under essentially identical conditions (or about as close as humanly possible), with a simple, defined, reproducible environment.
Further Reading
Parallel evolution
- Lenski, R. E., and M. Travisano. 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA 91: 6808-6814.
- Cooper, V. S., D. Schneider, M. Blot, and R. E. Lenski. 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183: 2834-2841.
- Cooper, T. F., D. E. Rozen, and R. E. Lenski. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 100: 1072-1077.
- Woods, R., D. Schneider, C. L. Winkworth, M. A. Riley, and R. E. Lenski. 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. USA 103: 9107-9112.
Divergent evolution
- Travisano, M., F. Vasi, and R. E. Lenski. 1995. Long-term experimental evolution in Escherichia coli. III. Variation among replicate populations in correlated responses to novel environments. Evolution 49: 189-200.
- Blount, Z. D., C. Z. Borland, and R. E. Lenski. 2008. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl. Acad. Sci. USA 105: 899-7906.
- Meyer, J. R., A. A. Agrawal, R. T. Quick, D. T. Dobias, D. Schneider, and R. E. Lenski. 2010. Parallel changes in host resistance to viral infection during 45,000 generations of relaxed selection. Evolution 64: 3024-3034.
- Blount, Z. D., J. E. Barrick, C. J. Davidson, and R. E. Lenski. 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489: 513-518.
