There is a quote by Theodosius Dobzhansky that goes: “Nothing in biology makes sense except in the light of evolution.” In response, I wish to offer up a corollary: “Nothing in evolution makes sense except in the light of molecular biology and genetics.”* Building a comprehensive model of evolution requires a thorough understanding of the molecular events involved. Here is where experimental evolution coupled with modern molecular biology and next-generation sequencing can create new ways for us to answer fundamental questions about the nature of evolution.
This summer, I had the pleasure of working at the Barrick Lab to understand some of the changes in the genomes of the 12 populations of the LTEE. Chromosomal rearrangements include insertions, deletions, duplications and inversions. While point mutations affect a single base, chromosomal rearrangements affect multiple genetic loci at once. These rearrangements can change chromosomal organization, alter gene copy number and expression, produce chimeric gene products, and affect mutation rates. In both wild microbial populations and other experimental evolution studies, chromosomal rearrangements have been shown to play a pivotal role in evolution.
Previously, the genomes of the LTEE populations were generally sequenced with short-read sequencing methods, which are good at looking for small mutations, but have trouble with resolving larger genomic structural variants. This problem is worsened by the fact that most rearrangements are mediated by repeat or transposable elements. This is where long-read sequencing methods, like Oxford Nanopore, come in handy. We sequenced clones from the 12 populations of the LTEE at 75,000 generations, and at additional time points for Ara+1 and Ara-3. Using a combination of computational tools, we generated genome assemblies and compared them to the ancestor REL606 to see what changes have occurred since the start of the experiment more than 3 decades ago.
Turns out, there are a lot of interesting things going on inside these humble 4,600,000-base-pair E. coli genomes. Almost all populations have deletions, while inversions and duplications were observed only in some populations. For example, Ara+4 has no inversions, while Ara+1 has the most extensive changes, with complex rearrangements that appear to be produced by successive inversions. Most of these rearrangements appear to be mediated by recombination among insertion sequence (IS) elements in the genome.
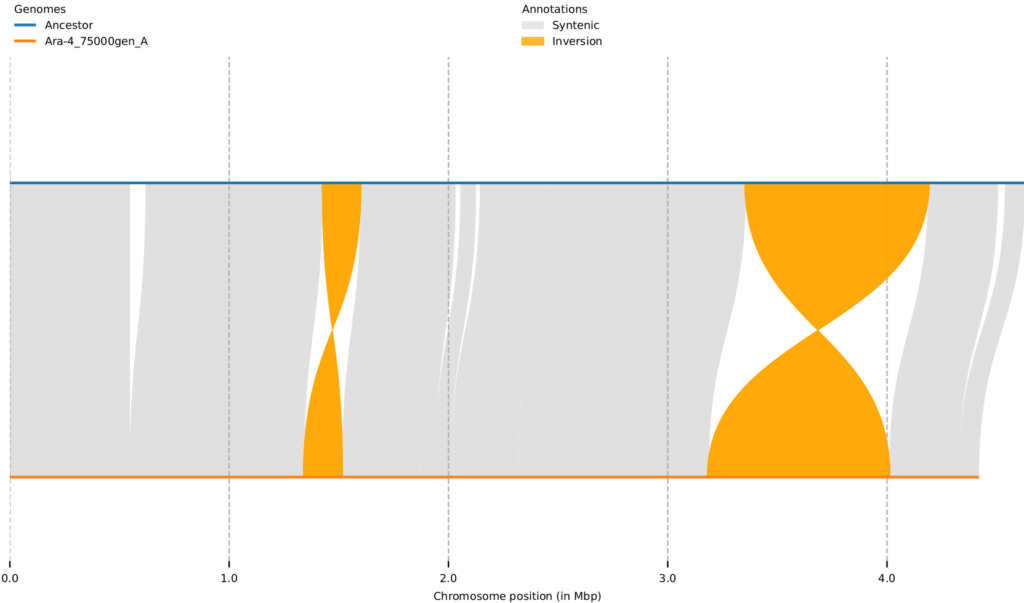
The obvious question to ask is are these rearrangements adaptive? Their continued existence in the LTEE would indicate that at the very least they are not deleterious. Some of these rearrangements are seen in two or more populations, and this parallelism can indicate adaptive benefit, or merely reflect a high rate of certain recombination events occurring.
Few experiments have captured my imagination like the LTEE has; it introduced me to an entirely new way to think about evolution. It exemplifies the kind of science I want to do: conceptually simple yet ambitious in its desire to answer open-ended questions about how life on earth evolves. On some days when I sit at my desk in the lab, I think about how just a few feet away, in 12 flasks kept warm in an incubator, endless forms most beautiful and most wonderful are evolving.

Ira Zibbu is pursuing her Master’s in Biology at the Indian Institute of Science Education and Research, Thiruvananthapuram. She can be reached at ira.zibbu@gmail.com. This work was support by the Khorana Scholars Program, which is funded by the Department of Biotechnology (DBT), Government of India, the Indo-U.S. Science and Technology Forum (IUSSTF) and WINStep Forward. It was also made possible with the support of the Department of Molecular Biosciences at the University of Texas at Austin
*The author wishes to declare a conflict of interest given that she works in molecular biology and genetics