2018
Blount Z D; Lenski R E; Losos J B
Contingency and determinism in evolution: Replaying life's tape Journal Article
Science, 362 (6415), pp. eaam5979, 2018, ISSN: 0036-8075.
Abstract | Links | BibTeX | Altmetric | Tags: Historical Contingency, Parallelism and Divergence, Review Articles
@article{Blount2018,
title = {Contingency and determinism in evolution: Replaying life's tape},
author = {Zachary D. Blount and Richard E. Lenski and Jonathan B. Losos},
url = {https://www.sciencemag.org/lookup/doi/10.1126/science.aam5979},
doi = {10.1126/science.aam5979},
issn = {0036-8075},
year = {2018},
date = {2018-11-01},
urldate = {2018-11-01},
journal = {Science},
volume = {362},
number = {6415},
pages = {eaam5979},
abstract = {Historical processes display some degree of "contingency," meaning their outcomes are sensitive to seemingly inconsequential events that can fundamentally change the future. Contingency is what makes historical outcomes unpredictable. Unlike many other natural phenomena, evolution is a historical process. Evolutionary change is often driven by the deterministic force of natural selection, but natural selection works upon variation that arises unpredictably through time by random mutation, and even beneficial mutations can be lost by chance through genetic drift. Moreover, evolution has taken place within a planetary environment with a particular history of its own. This tension between determinism and contingency makes evolutionary biology a kind of hybrid between science and history. While philosophers of science examine the nuances of contingency, biologists have performed many empirical studies of evolutionary repeatability and contingency. Here, we review the experimental and comparative evidence from these studies. Replicate populations in evolutionary "replay" experiments often show parallel changes, especially in overall performance, although idiosyncratic outcomes show that the particulars of a lineage's history can affect which of several evolutionary paths is taken. Comparative biologists have found many notable examples of convergent adaptation to similar conditions, but quantification of how frequently such convergence occurs is difficult. On balance, the evidence indicates that evolution tends to be surprisingly repeatable among closely related lineages, but disparate outcomes become more likely as the footprint of history grows deeper. Ongoing research on the structure of adaptive landscapes is providing additional insight into the interplay of fate and chance in the evolutionary process.},
keywords = {Historical Contingency, Parallelism and Divergence, Review Articles},
pubstate = {published},
tppubtype = {article}
}
Bajić D; Vila J C C; Blount Z D; Sánchez A
On the deformability of an empirical fitness landscape by microbial evolution Journal Article
Proceedings of the National Academy of Sciences of the United States of America, 115 (44), pp. 11286-11291, 2018, ISSN: 0027-8424.
Abstract | Links | BibTeX | Altmetric | Tags: Citrate Evolution, Demography and Ecology, Theory and Simulations
@article{nokey,
title = {On the deformability of an empirical fitness landscape by microbial evolution},
author = {Djordje Bajić and Jean C. C. Vila and Zachary D. Blount and Alvaro Sánchez},
url = {http://www.pnas.org/lookup/doi/10.1073/pnas.1808485115},
doi = {10.1073/pnas.1808485115},
issn = {0027-8424},
year = {2018},
date = {2018-10-30},
urldate = {2018-10-30},
journal = {Proceedings of the National Academy of Sciences of the United States of America},
volume = {115},
number = {44},
pages = {11286-11291},
abstract = {A fitness landscape is a map between the genotype and its reproductive success in a given environment. The topography of fitness landscapes largely governs adaptive dynamics, constraining evolutionary trajectories and the predictability of evolution. Theory suggests that this topography can be deformed by mutations that produce substantial changes to the environment. Despite its importance, the deformability of fitness landscapes has not been systematically studied beyond abstract models, and little is known about its reach and consequences in empirical systems. Here we have systematically characterized the deformability of the genome-wide metabolic fitness landscape of the bacterium \textit{Escherichia coli}. Deformability is quantified by the noncommutativity of epistatic interactions, which we experimentally demonstrate in mutant strains on the path to an evolutionary innovation. Our analysis shows that the deformation of fitness landscapes by metabolic mutations rarely affects evolutionary trajectories in the short range. However, mutations with large environmental effects produce long-range landscape deformations in distant regions of the genotype space that affect the fitness of later descendants. Our results therefore suggest that, even in situations in which mutations have strong environmental effects, fitness landscapes may retain their power to forecast evolution over small mutational distances despite the potential attenuation of that power over longer evolutionary trajectories. Our methods and results provide an avenue for integrating adaptive and eco-evolutionary dynamics with complex genetics and genomics. },
keywords = {Citrate Evolution, Demography and Ecology, Theory and Simulations},
pubstate = {published},
tppubtype = {article}
}
Leon D; D'Alton S; Quandt E M; Barrick J E
Innovation in an E. coli evolution experiment is contingent on maintaining adaptive potential until competition subsides Journal Article
PLOS Genetics, 14 (4), pp. e1007348, 2018, ISSN: 1553-7404.
Abstract | Links | BibTeX | Altmetric | Tags: Citrate Evolution, Demography and Ecology, Historical Contingency
@article{nokey,
title = {Innovation in an \textit{E. coli} evolution experiment is contingent on maintaining adaptive potential until competition subsides},
author = {Dacia Leon and Simon D'Alton and Erik M. Quandt and Jeffrey E. Barrick},
url = {https://dx.plos.org/10.1371/journal.pgen.1007348},
doi = {10.1371/journal.pgen.1007348},
issn = {1553-7404},
year = {2018},
date = {2018-04-12},
urldate = {2018-04-12},
journal = {PLOS Genetics},
volume = {14},
number = {4},
pages = {e1007348},
abstract = {Key innovations are disruptive evolutionary events that enable a species to escape constraints and rapidly diversify. After 15 years of the Lenski long-term evolution experiment with \textit{Escherichia coli}, cells in one of the twelve populations evolved the ability to utilize citrate, an abundant but previously untapped carbon source in the environment. Descendants of these cells became dominant in the population and subsequently diversified as a consequence of invading this vacant niche. Mutations responsible for the appearance of rudimentary citrate utilization and for refining this ability have been characterized. However, the complete nature of the genetic and/or ecological events that set the stage for this key innovation is unknown. In particular, it is unclear why it took so long for citrate utilization to evolve and why it still has evolved in only one of the twelve \textit{E. coli} populations after 30 years of the Lenski experiment. In this study, we recapitulated the initial mutation needed to evolve citrate utilization in strains isolated from throughout the first 31,500 generations of the history of this population. We found that there was already a slight fitness benefit for this mutation in the original ancestor of the evolution experiment and in other early isolates. However, evolution of citrate utilization was blocked at this point due to competition with other mutations that improved fitness in the original niche. Subsequently, an anti-potentiated genetic background evolved in which it was deleterious to evolve rudimentary citrate utilization. Only later, after further mutations accumulated that restored the benefit of this first-step mutation and the overall rate of adaptation in the population slowed, was citrate utilization likely to evolve. Thus, intense competition and the types of mutations that it favors can lead to short-sighted evolutionary trajectories that hide a stepping stone needed to access a key innovation from many future generations.},
keywords = {Citrate Evolution, Demography and Ecology, Historical Contingency},
pubstate = {published},
tppubtype = {article}
}
Lenski R E; Burnham T C
Experimental evolution of bacteria across 60,000 generations, and what it might mean for economics and human decision-making Journal Article
Journal of Bioeconomics, 20 (1), pp. 107–124, 2018, ISSN: 1387-6996.
Abstract | Links | BibTeX | Altmetric | Tags: Methods and Miscellaneous
@article{Lenski2018,
title = {Experimental evolution of bacteria across 60,000 generations, and what it might mean for economics and human decision-making},
author = {Richard E. Lenski and Terence C. Burnham},
url = {http://link.springer.com/10.1007/s10818-017-9258-7},
doi = {10.1007/s10818-017-9258-7},
issn = {1387-6996},
year = {2018},
date = {2018-04-01},
urldate = {2018-04-01},
journal = {Journal of Bioeconomics},
volume = {20},
number = {1},
pages = {107--124},
publisher = {Springer US},
abstract = {Evolutionary biology and economics are both rich in theory and steeped in data, but they also share challenges including the fact that the systems they seek to understand are, in certain respects, unique and not easily manipulated. Nonetheless, both fields have seen growing efforts to provide experimental approaches to address specific issues. Here, we review some results from a 30-year experiment in which 12 populations of bacteria have been evolving for over 60,000 generations to characterize: (i) the time scale of adaptation to new conditions, (ii) the repeatability of evolutionary changes, and (iii) the benefits and costs of specialization. In each case, we speculate on potential connections and implications of these findings for the field of economics. Moreover, both the bacteria in this experiment and people in modern societies live in novel environments, which leads to an evolutionary mismatch between their genes and environments. Regardless of the value of our speculations, we hope this paper stimulates further interest in pursuing experiments in fields that are often viewed as observational and not amenable to experimentation.},
keywords = {Methods and Miscellaneous},
pubstate = {published},
tppubtype = {article}
}
Peng F; Widmann S; Wünsche A; Duan K; Donovan K A; Dobson R C J; Lenski R E; Cooper T F
Effects of Beneficial Mutations in pykF Gene Vary over Time and across Replicate Populations in a Long-Term Experiment with Bacteria Journal Article
Molecular Biology and Evolution, 35 (1), pp. 202–210, 2018, ISSN: 0737-4038.
Abstract | Links | BibTeX | Altmetric | Tags: Genotypes and Phenotypes, Historical Contingency
@article{Peng2018,
title = {Effects of Beneficial Mutations in \textit{pykF} Gene Vary over Time and across Replicate Populations in a Long-Term Experiment with Bacteria},
author = {Fen Peng and Scott Widmann and Andrea Wünsche and Kristina Duan and Katherine A. Donovan and Renwick C J Dobson and Richard E. Lenski and Tim F. Cooper},
url = {https://academic.oup.com/mbe/article/35/1/202/4562833},
doi = {10.1093/molbev/msx279},
issn = {0737-4038},
year = {2018},
date = {2018-01-01},
urldate = {2018-01-01},
journal = {Molecular Biology and Evolution},
volume = {35},
number = {1},
pages = {202--210},
abstract = {The fitness effects of mutations can depend on the genetic backgrounds in which they occur and thereby influence future opportunities for evolving populations. In particular, mutations that fix in a population might change the selective benefit of subsequent mutations, giving rise to historical contingency. We examine these effects by focusing on mutations in a key metabolic gene, \textit{pykF}, that arose independently early in the history of 12 \textit{Escherichia coli} populations during a long-Term evolution experiment. Eight different evolved nonsynonymous mutations conferred similar fitness benefits of ~10% when transferred into the ancestor, and these benefits were greater than the one conferred by a deletion mutation. In contrast, the same mutations had highly variable fitness effects, ranging from ~0% to 25%, in evolved clones isolated from the populations at 20,000 generations. Two mutations that were moved into these evolved clones conferred similar fitness effects in a given clone, but different effects between the clones, indicating epistatic interactions between the evolved \textit{pykF} alleles and the other mutations that had accumulated in each evolved clone. We also measured the fitness effects of six evolved \textit{pykF} alleles in the same populations in which they had fixed, but at seven time points between 0 and 50,000 generations. Variation in fitness effects was high at intermediate time points, and declined to a low level at 50,000 generations, when the mean fitness effect was lowest. Our results demonstrate the importance of genetic context in determining the fitness effects of different beneficial mutations even within the same gene.},
keywords = {Genotypes and Phenotypes, Historical Contingency},
pubstate = {published},
tppubtype = {article}
}
Maddamsetti R; Lenski R E
PLOS Genetics, 14 (1), pp. e1007199, 2018, ISSN: 1553-7404.
Abstract | Links | BibTeX | Altmetric | Tags: Descendant Experiments, Genome Evolution
@article{Maddamsetti2018,
title = {Analysis of bacterial genomes from an evolution experiment with horizontal gene transfer shows that recombination can sometimes overwhelm selection},
author = {Rohan Maddamsetti and Richard E. Lenski},
editor = {Ivan Matic},
url = {https://dx.plos.org/10.1371/journal.pgen.1007199},
doi = {10.1371/journal.pgen.1007199},
issn = {1553-7404},
year = {2018},
date = {2018-01-01},
urldate = {2018-01-01},
journal = {PLOS Genetics},
volume = {14},
number = {1},
pages = {e1007199},
abstract = {Few experimental studies have examined the role that sexual recombination plays in bacterial evolution, including the effects of horizontal gene transfer on genome structure. To address this limitation, we analyzed genomes from an experiment in which \textit{Escherichia coli} K-12 Hfr (high frequency recombination) donors were periodically introduced into 12 evolving populations of \textit{E. coli} B and allowed to conjugate repeatedly over the course of 1000 generations. Previous analyses of the evolved strains from this experiment showed that recombination did not accelerate adaptation, despite increasing genetic variation relative to asexual controls. However, the resolution in that previous work was limited to only a few genetic markers. We sought to clarify and understand these puzzling results by sequencing complete genomes from each population. The effects of recombination were highly variable: one lineage was mostly derived from the donors, while another acquired almost no donor DNA. In most lineages, some regions showed repeated introgression and others almost none. Regions with high introgression tended to be near the donors' origin of transfer sites. To determine whether introgressed alleles imposed a genetic load, we extended the experiment for 200 generations without recombination and sequenced whole-population samples. Beneficial alleles in the recipient populations were occasionally driven extinct by maladaptive donor-derived alleles. On balance, our analyses indicate that the plasmid-mediated recombination was sufficiently frequent to drive donor alleles to fixation without providing much, if any, selective advantage.},
keywords = {Descendant Experiments, Genome Evolution},
pubstate = {published},
tppubtype = {article}
}
2017
Lenski R E
What is adaptation by natural selection? Perspectives of an experimental microbiologist Journal Article
PLOS Genetics, 13 (4), pp. e1006668, 2017, ISSN: 1553-7404.
Abstract | Links | BibTeX | Altmetric | Tags: Review Articles
@article{Lenski2017,
title = {What is adaptation by natural selection? Perspectives of an experimental microbiologist},
author = {Richard E. Lenski},
editor = {W. Ford Doolittle},
url = {https://dx.plos.org/10.1371/journal.pgen.1006668},
doi = {10.1371/journal.pgen.1006668},
issn = {1553-7404},
year = {2017},
date = {2017-04-01},
urldate = {2017-04-01},
journal = {PLOS Genetics},
volume = {13},
number = {4},
pages = {e1006668},
abstract = {Use of at least three potent antiretroviral agents has become the standard of care in the management of HIV infection. The potential toxicities associated with highly active antiretroviral therapy (HAART) however, may limit a patient's ability to adhere to and tolerate these agents. Although a comprehensive discussion of all toxicities associated with HAART is beyond the scope of this article, selected short-term and long-term significant toxicities will be reviewed. Short-term toxicities that will be discussed include abacavir-induced hypersensitivity reactions, efavirenz-associated central nervous system side effects and rash associated with the non-nucleoside reverse transcriptase inhibitors (NNRTIs) and the protease inhibitor (PI) amprenavir. Several long-term toxicities associated with the nucleoside reverse transcriptase inhibitors (NRTIs) are hypothesized to be due to mitochondrial toxicity. These toxicities include myositis and lactic acidosis with hepatic steatosis, pancreatitis and peripheral neuropathy. Some experts also hypothesize that mitochondrial toxicity is responsible for the lipodystrophy syndrome, which includes hyperglycemia, abnormal fat redistribution and dyslipidemia. Finally, indinavir-associated nephrolithiasis, which may present with either short term or long term use will be discussed. This article will provide the practicing pharmacist with a review of these significant toxicities, the implicated agents, incidence, usual clinical presentation, and recommendations for management.},
keywords = {Review Articles},
pubstate = {published},
tppubtype = {article}
}
Deatherage D E; Kepner J L; Bennett A F; Lenski R E; Barrick J E
Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures Journal Article
Proceedings of the National Academy of Sciences of the United States of America, 114 (10), pp. E1904–E1912, 2017, ISSN: 0027-8424.
Abstract | Links | BibTeX | Altmetric | Tags: Descendant Experiments
@article{Deatherage2017,
title = {Specificity of genome evolution in experimental populations of \textit{Escherichia coli} evolved at different temperatures},
author = {Daniel E. Deatherage and Jamie L. Kepner and Albert F. Bennett and Richard E. Lenski and Jeffrey E. Barrick},
url = {http://www.pnas.org/lookup/doi/10.1073/pnas.1616132114},
doi = {10.1073/pnas.1616132114},
issn = {0027-8424},
year = {2017},
date = {2017-03-01},
urldate = {2017-03-01},
journal = {Proceedings of the National Academy of Sciences of the United States of America},
volume = {114},
number = {10},
pages = {E1904--E1912},
abstract = {Isolated populations derived from a common ancestor are expected to diverge genetically and phenotypically as they adapt to different local environments. To examine this process, 30 populations of \textit{Escherichia coli} were evolved for 2,000 generations, with six in each of five different thermal regimes: constant 20 °C, 32 °C, 37 °C, 42 °C, and daily alternations between 32 °C and 42 °C. Here, we sequenced the genomes of one endpoint clone from each population to test whether the history of adaptation in different thermal regimes was evident at the genomic level. The evolved strains had accumulated ∼5.3 mutations, on average, and exhibited distinct signatures of adaptation to the different environments. On average, two strains that evolved under the same regime exhibited ∼17% overlap in which genes were mutated, whereas pairs that evolved under different conditions shared only ∼4%. For example, all six strains evolved at 32 °C had mutations in nadR , whereas none of the other 24 strains did. However, a population evolved at 37 °C for an additional 18,000 generations eventually accumulated mutations in the signature genes strongly associated with adaptation to the other temperature regimes. Two mutations that arose in one temperature treatment tended to be beneficial when tested in the others, although less so than in the regime in which they evolved. These findings demonstrate that genomic signatures of adaptation can be highly specific, even with respect to subtle environmental differences, but that this imprint may become obscured over longer timescales as populations continue to change and adapt to the shared features of their environments.},
keywords = {Descendant Experiments},
pubstate = {published},
tppubtype = {article}
}
Good B H; McDonald M J; Barrick J E; Lenski R E; Desai M M
The dynamics of molecular evolution over 60,000 generations Journal Article
Nature, 551 (7678), pp. 45–50, 2017, ISSN: 14764687.
Abstract | Links | BibTeX | Altmetric | Tags: Demography and Ecology, Genome Evolution, Historical Contingency, Mutation Rates, Parallelism and Divergence
@article{Good2017,
title = {The dynamics of molecular evolution over 60,000 generations},
author = {Benjamin H. Good and Michael J. McDonald and Jeffrey E. Barrick and Richard E. Lenski and Michael M. Desai},
url = {http://dx.doi.org/10.1038/nature24287},
doi = {10.1038/nature24287},
issn = {14764687},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {Nature},
volume = {551},
number = {7678},
pages = {45--50},
publisher = {Nature Publishing Group},
abstract = {The outcomes of evolution are determined by a stochastic dynamical process that governs how mutations arise and spread through a population. However, it is difficult to observe these dynamics directly over long periods and across entire genomes. Here we analyse the dynamics of molecular evolution in twelve experimental populations of \textit{Escherichia coli}, using whole-genome metagenomic sequencing at five hundred-generation intervals through sixty thousand generations. Although the rate of fitness gain declines over time, molecular evolution is characterized by signatures of rapid adaptation throughout the duration of the experiment, with multiple beneficial variants simultaneously competing for dominance in each population. Interactions between ecological and evolutionary processes play an important role, as long-term quasi-stable coexistence arises spontaneously in most populations, and evolution continues within each clade. We also present evidence that the targets of natural selection change over time, as epistasis and historical contingency alter the strength of selection on different genes. Together, these results show that long-term adaptation to a constant environment can be a more complex and dynamic process than is often assumed.},
keywords = {Demography and Ecology, Genome Evolution, Historical Contingency, Mutation Rates, Parallelism and Divergence},
pubstate = {published},
tppubtype = {article}
}
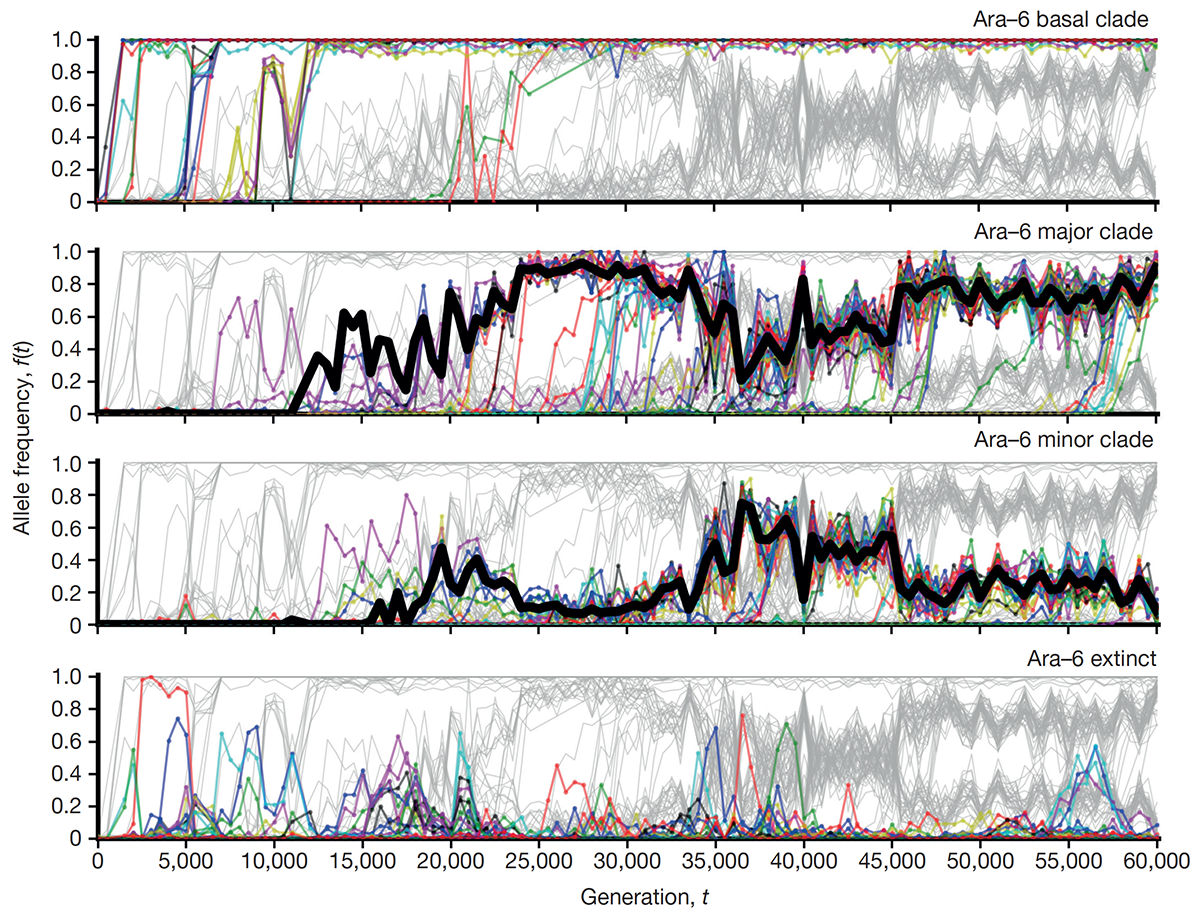
Maddamsetti R; Hatcher P J; Green A G; Williams B L; Marks D S; Lenski R E
Core genes evolve rapidly in the long-term evolution experiment with Escherichia coli Journal Article
Genome Biology and Evolution, 9 (4), pp. 1072–1083, 2017, ISSN: 17596653.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution, Genotypes and Phenotypes, Mutation Rates
@article{Maddamsetti2017,
title = {Core genes evolve rapidly in the long-term evolution experiment with \textit{Escherichia coli}},
author = {Rohan Maddamsetti and Philip J. Hatcher and Anna G. Green and Barry L. Williams and Debora S. Marks and Richard E. Lenski},
url = {https://academic.oup.com/gbe/article/9/4/1072/3100447},
doi = {10.1093/gbe/evx064},
issn = {17596653},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {Genome Biology and Evolution},
volume = {9},
number = {4},
pages = {1072--1083},
abstract = {Bacteria can evolve rapidly under positive selection owing to their vast numbers, allowing their genes to diversify by adapting to different environments. We asked whether the same genes that evolve rapidly in the long-term evolution experiment (LTEE) with \textit{Escherichia coli} have also diversified extensively in nature. To make this comparison, we identified ~2000 core genes shared among 60 \textit{E. coli} strains. During the LTEE, core genes accumulated significantly more nonsynonymous mutations than flexible (i.e., noncore) genes. Furthermore, core genes under positive selection in the LTEE are more conserved in nature than the average core gene. In some cases, adaptive mutations appear to modify protein functions, rather than merely knocking them out. The LTEE conditions are novel for \textit{E. coli}, at least in relation to its evolutionary history in nature. The constancy and simplicity of the environment likely favor the complete loss of some unused functions and the fine-tuning of others.},
keywords = {Genome Evolution, Genotypes and Phenotypes, Mutation Rates},
pubstate = {published},
tppubtype = {article}
}
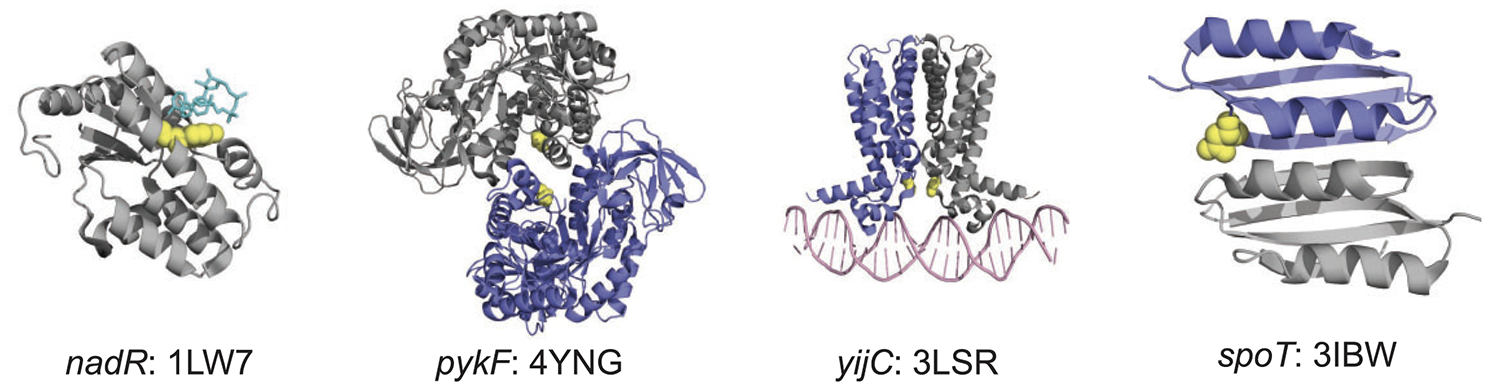
2016
Großkopf T; Consuegra J; Gaffé J; Willison J C; Lenski R E; Soyer O S; Schneider D
Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment Journal Article
BMC Evolutionary Biology, 16 (1), pp. 163, 2016, ISSN: 1471-2148.
Abstract | Links | BibTeX | Altmetric | Tags: Demography and Ecology, Theory and Simulations
@article{Großkopf2016,
title = {Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment},
author = {Tobias Großkopf and Jessika Consuegra and Joël Gaffé and John C. Willison and Richard E. Lenski and Orkun S. Soyer and Dominique Schneider},
url = {https://bmcevolbiol.biomedcentral.com/articles/10.1186/s12862-016-0733-x},
doi = {10.1186/s12862-016-0733-x},
issn = {1471-2148},
year = {2016},
date = {2016-12-01},
urldate = {2016-12-01},
journal = {BMC Evolutionary Biology},
volume = {16},
number = {1},
pages = {163},
publisher = {BMC Evolutionary Biology},
abstract = {Background
Predicting adaptive trajectories is a major goal of evolutionary biology and useful for practical applications. Systems biology has enabled the development of genome-scale metabolic models. However, analysing these models via flux balance analysis (FBA) cannot predict many evolutionary outcomes including adaptive diversification, whereby an ancestral lineage diverges to fill multiple niches. Here we combine in silico evolution with FBA and apply this modelling framework, evoFBA, to a long-term evolution experiment with \textit{Escherichia coli}.
Results
Simulations predicted the adaptive diversification that occurred in one experimental population and generated hypotheses about the mechanisms that promoted coexistence of the diverged lineages. We experimentally tested and, on balance, verified these mechanisms, showing that diversification involved niche construction and character displacement through differential nutrient uptake and altered metabolic regulation.
Conclusion
The evoFBA framework represents a promising new way to model biochemical evolution, one that can generate testable predictions about evolutionary and ecosystem-level outcomes.},
keywords = {Demography and Ecology, Theory and Simulations},
pubstate = {published},
tppubtype = {article}
}
Predicting adaptive trajectories is a major goal of evolutionary biology and useful for practical applications. Systems biology has enabled the development of genome-scale metabolic models. However, analysing these models via flux balance analysis (FBA) cannot predict many evolutionary outcomes including adaptive diversification, whereby an ancestral lineage diverges to fill multiple niches. Here we combine in silico evolution with FBA and apply this modelling framework, evoFBA, to a long-term evolution experiment with Escherichia coli.
Results
Simulations predicted the adaptive diversification that occurred in one experimental population and generated hypotheses about the mechanisms that promoted coexistence of the diverged lineages. We experimentally tested and, on balance, verified these mechanisms, showing that diversification involved niche construction and character displacement through differential nutrient uptake and altered metabolic regulation.
Conclusion
The evoFBA framework represents a promising new way to model biochemical evolution, one that can generate testable predictions about evolutionary and ecosystem-level outcomes.
Tenaillon O; Barrick J E; Ribeck N; Deatherage D E; Blanchard J L; Dasgupta A; Wu G C; Wielgoss S; Cruveiller S; Medigue C; Schneider D; Lenski R E
Tempo and mode of genome evolution in a 50,000-generation experiment. Journal Article
Nature, 536 (7615), pp. 165–170, 2016, ISSN: 1476-4687.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution, Mutation Rates, Parallelism and Divergence
@article{Tenaillon2016,
title = {Tempo and mode of genome evolution in a 50,000-generation experiment.},
author = {Olivier Tenaillon and Jeffrey E. Barrick and Noah Ribeck and Daniel E. Deatherage and Jeffrey L. Blanchard and Aurko Dasgupta and Gabriel C. Wu and Sebastien Wielgoss and Stephane Cruveiller and Claudine Medigue and Dominique Schneider and Richard E. Lenski},
url = {http://www.ncbi.nlm.nih.gov/pubmed/27479321},
doi = {10.1038/nature18959},
issn = {1476-4687},
year = {2016},
date = {2016-08-01},
urldate = {2016-08-01},
journal = {Nature},
volume = {536},
number = {7615},
pages = {165--170},
publisher = {Nature Publishing Group},
abstract = {Adaptation by natural selection depends on the rates, effects and interactions of many mutations, making it difficult to determine what proportion of mutations in an evolving lineage are beneficial. Here we analysed 264 complete genomes from 12 \textit{Escherichia coli} populations to characterize their dynamics over 50,000 generations. The populations that retained the ancestral mutation rate support a model in which most fixed mutations are beneficial, the fraction of beneficial mutations declines as fitness rises, and neutral mutations accumulate at a constant rate. We also compared these populations to mutation-accumulation lines evolved under a bottlenecking regime that minimizes selection. Nonsynonymous mutations, intergenic mutations, insertions and deletions are overrepresented in the long-term populations, further supporting the inference that most mutations that reached high frequency were favoured by selection. These results illuminate the shifting balance of forces that govern genome evolution in populations adapting to a new environment.},
keywords = {Genome Evolution, Mutation Rates, Parallelism and Divergence},
pubstate = {published},
tppubtype = {article}
}
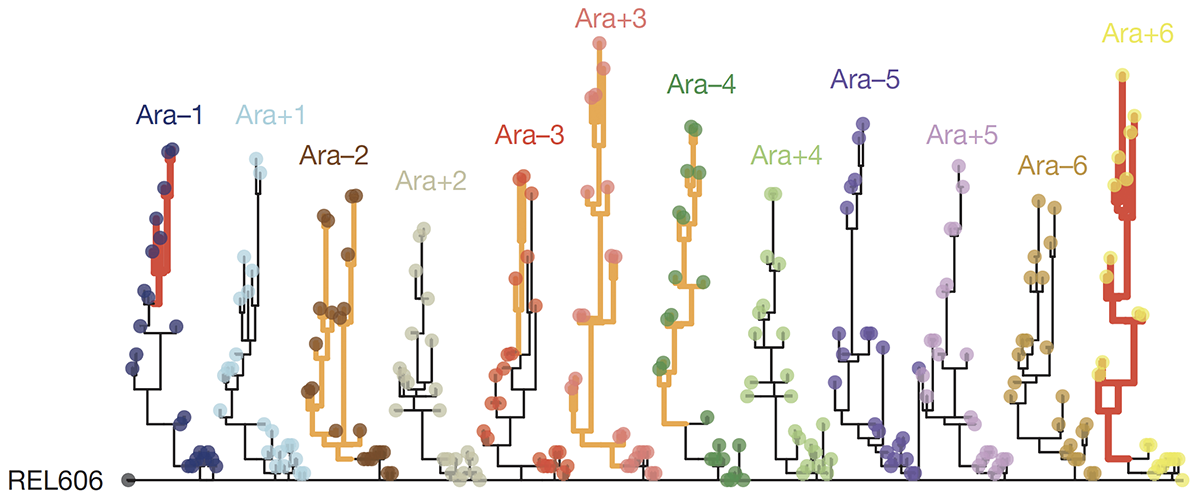
Blount Z D
A case study in evolutionary contingency Journal Article
Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences, 58 , pp. 82–92, 2016, ISSN: 13698486.
Abstract | Links | BibTeX | Altmetric | Tags: Review Articles
@article{Blount2016,
title = {A case study in evolutionary contingency},
author = {Zachary D. Blount},
url = {https://linkinghub.elsevier.com/retrieve/pii/S1369848615001806},
doi = {10.1016/j.shpsc.2015.12.007},
issn = {13698486},
year = {2016},
date = {2016-08-01},
urldate = {2016-08-01},
journal = {Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences},
volume = {58},
pages = {82--92},
publisher = {Elsevier Ltd},
abstract = {Biological evolution is a fundamentally historical phenomenon in which intertwined stochastic and deterministic processes shape lineages with long, continuous histories that exist in a changing world that has a history of its own. The degree to which these characteristics render evolution historically contingent, and evolutionary outcomes thereby unpredictably sensitive to history has been the subject of considerable debate in recent decades. Microbial evolution experiments have proven among the most fruitful means of empirically investigating the issue of historical contingency in evolution. One such experiment is the \textit{Escherichia coli} Long-Term Evolution Experiment (LTEE), in which twelve populations founded from the same clone of \textit{E. coli} have evolved in parallel under identical conditions. Aerobic growth on citrate (Cit+), a novel trait for \textit{E. coli}, evolved in one of these populations after more than 30,000 generations. Experimental replays of this population's evolution from various points in its history showed that the Cit+ trait was historically contingent upon earlier mutations that potentiated the trait by rendering it mutationally accessible. Here I review this case of evolutionary contingency and discuss what it implies about the importance of historical contingency arising from the core processes of evolution.},
keywords = {Review Articles},
pubstate = {published},
tppubtype = {article}
}
Ribeck N; Mulka J S; Zaman L; Connelly B D; Lenski R E
Competition between continuously evolving lineages in asexual populations Journal Article
bioRxiv, 2016.
Abstract | Links | BibTeX | Altmetric | Tags: Theory and Simulations
@article{Ribeck062976,
title = {Competition between continuously evolving lineages in asexual populations},
author = {Noah Ribeck and Joseph S. Mulka and Luis Zaman and Brian D. Connelly and Richard E. Lenski},
url = {https://www.biorxiv.org/content/early/2016/07/10/062976},
doi = {10.1101/062976},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
journal = {bioRxiv},
publisher = {Cold Spring Harbor Laboratory},
abstract = {In an asexual population, the fate of a beneficial mutation depends on how its lineage competes against other mutant lineages in the population. With high beneficial mutation rates or large population sizes, competition between contending mutations is strong, and successful lineages can accumulate multiple mutations before any single one achieves fixation. Most current theory about asexual population dynamics either neglects this multiple-mutations regime or introduces simplifying assumptions that may not apply. Here, we develop a theoretical framework that describes the dynamics of adaptation and substitution over all mutation-rate regimes by conceptualizing the population as a collection of continuously adapting lineages. This model of textquotedblleftlineage interferencetextquotedblright shows that each new mutanttextquoterights advantage over the rest of the population must be above a critical threshold in order to likely achieve fixation, and we derive a simple expression for that threshold. We apply this framework to examine the role of beneficial mutations with different effect sizes across the transition to the multiple-mutations regime.},
keywords = {Theory and Simulations},
pubstate = {published},
tppubtype = {article}
}
2015
Lenski R E; Wiser M J; Ribeck N; Blount Z D; Nahum J R; Morris J J; Zaman L; Turner C B; Wade B D; Maddamsetti R; Burmeister A R; Baird E J; Bundy J; Grant N A; Card K J; Rowles M; Weatherspoon K; Papoulis S E; Sullivan R; Clark C; Mulka J S; Hajela N
Sustained fitness gains and variability in fitness trajectories in the long-term evolution experiment with Escherichia coli Journal Article
Proceedings of the Royal Society B: Biological Sciences, 282 (1821), pp. 20152292, 2015, ISSN: 0962-8452.
Abstract | Links | BibTeX | Altmetric | Tags: Fitness Trajectories, Mutation Rates, Parallelism and Divergence
@article{nokey,
title = {Sustained fitness gains and variability in fitness trajectories in the long-term evolution experiment with \textit{Escherichia coli}},
author = {Richard E. Lenski and Michael J. Wiser and Noah Ribeck and Zachary D. Blount and Joshua R. Nahum and J. Jeffrey Morris and Luis Zaman and Caroline B. Turner and Brian D. Wade and Rohan Maddamsetti and Alita R. Burmeister and Elizabeth J. Baird and Jay Bundy and Nkrumah A. Grant and Kyle J. Card and Maia Rowles and Kiyana Weatherspoon and Spiridon E. Papoulis and Rachel Sullivan and Colleen Clark and Joseph S. Mulka and Neerja Hajela},
url = {https://royalsocietypublishing.org/doi/10.1098/rspb.2015.2292},
doi = {10.1098/rspb.2015.2292},
issn = {0962-8452},
year = {2015},
date = {2015-12-22},
urldate = {2015-12-22},
journal = {Proceedings of the Royal Society B: Biological Sciences},
volume = {282},
number = {1821},
pages = {20152292},
abstract = {Many populations live in environments subject to frequent biotic and abiotic changes. Nonetheless, it is interesting to ask whether an evolving population's mean fitness can increase indefinitely, and potentially without any limit, even in a constant environment. A recent study showed that fitness trajectories of \textit{Escherichia coli} populations over 50 000 generations were better described by a power-law model than by a hyperbolic model. According to the power-law model, the rate of fitness gain declines over time but fitness has no upper limit, whereas the hyperbolic model implies a hard limit. Here, we examine whether the previously estimated power-law model predicts the fitness trajectory for an additional 10 000 generations. To that end, we conducted more than 1100 new competitive fitness assays. Consistent with the previous study, the power-law model fits the new data better than the hyperbolic model. We also analysed the variability in fitness among populations, finding subtle, but significant, heterogeneity in mean fitness. Some, but not all, of this variation reflects differences in mutation rate that evolved over time. Taken together, our results imply that both adaptation and divergence can continue indefinitely—or at least for a long time—even in a constant environment.},
keywords = {Fitness Trajectories, Mutation Rates, Parallelism and Divergence},
pubstate = {published},
tppubtype = {article}
}
Maddamsetti R; Hatcher P J; Cruveiller S; Medigue C; Barrick J E; Lenski R E
Molecular Biology and Evolution, 32 (11), 2015, ISSN: 0737-4038.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution
@article{Maddamsetti2015,
title = {Synonymous Genetic Variation in Natural Isolates of \textit{Escherichia coli} Does Not Predict Where Synonymous Substitutions Occur in a Long-Term Experiment},
author = {Rohan Maddamsetti and Philip J. Hatcher and Stephane Cruveiller and Claudine Medigue and Jeffrey E. Barrick and Richard E. Lenski},
url = {https://academic.oup.com/mbe/article-lookup/doi/10.1093/molbev/msv161},
doi = {10.1093/molbev/msv161},
issn = {0737-4038},
year = {2015},
date = {2015-11-01},
urldate = {2015-11-01},
journal = {Molecular Biology and Evolution},
volume = {32},
number = {11},
abstract = {Synonymous genetic differences vary by more than 20-fold among genes in natural isolates of \textit{Escherichia coli}. One hypothesis to explain this heterogeneity is that genes with high levels of synonymous variation mutate at higher rates than genes with low synonymous variation. If so, then one would expect to observe similar mutational patterns in evolution experiments. In fact, however, the pattern of synonymous substitutions in a long-term evolution experiment with \textit{E. coli} does not support this hypothesis. In particular, the extent of synonymous variation across genes in that experiment does not reflect the variation observed in natural isolates of \textit{E. coli}. Instead, gene length alone predicts with high accuracy the prevalence of synonymous changes in the experimental populations. We hypothesize that patterns of synonymous variation in natural \textit{E. coli} populations are instead caused by differences across genomic regions in their effective population size that, in turn, reflect different histories of recombination, horizontal gene transfer, selection, and population structure.},
keywords = {Genome Evolution},
pubstate = {published},
tppubtype = {article}
}
Turner C B; Blount Z D; Lenski R E
Replaying Evolution to Test the Cause of Extinction of One Ecotype in an Experimentally Evolved Population Journal Article
PLOS ONE, 10 (11), pp. e0142050, 2015, ISSN: 1932-6203.
Abstract | Links | BibTeX | Altmetric | Tags: Citrate Evolution, Demography and Ecology, Historical Contingency
@article{Turner2015,
title = {Replaying Evolution to Test the Cause of Extinction of One Ecotype in an Experimentally Evolved Population},
author = {Caroline B. Turner and Zachary D. Blount and Richard E. Lenski},
editor = {Frederick M. Cohan},
url = {https://dx.plos.org/10.1371/journal.pone.0142050},
doi = {10.1371/journal.pone.0142050},
issn = {1932-6203},
year = {2015},
date = {2015-11-01},
urldate = {2015-11-01},
journal = {PLOS ONE},
volume = {10},
number = {11},
pages = {e0142050},
abstract = {In a long-term evolution experiment with \textit{Escherichia coli}, bacteria in one of twelve populations evolved the ability to consume citrate, a previously unexploited resource in a glucoselimited medium. This innovation led to the frequency-dependent coexistence of citrate-consuming (Cit+) and non-consuming (Cit-) ecotypes, with Cit-bacteria persisting on the exogenously supplied glucose as well as other carbon molecules released by the Cit+ bacteria. After more than 10,000 generations of coexistence, however, the Cit-lineage went extinct; cells with the Cit-phenotype dropped to levels below detection, and the Cit-clade could not be detected by molecular assays based on its unique genotype. We hypothesized that this extinction was a deterministic outcome of evolutionary change within the population, specifically the appearance of a more-fit Cit+ ecotype that competitively excluded the Cit-ecotype. We tested this hypothesis by re-evolving the population from a frozen population sample taken within 500 generations of the extinction and from another sample taken several thousand generations earlier, in each case for 500 generations and with 20-fold replication. To our surprise, the Cit-type did not go extinct in any of these replays, and Cit-cells also persisted in a single replicate that was propagated for 2,500 generations. Even more unexpectedly, we showed that the Cit-ecotype could reinvade the Cit+ population after its extinction. Taken together, these results indicate that the extinction of the Cit-ecotype was not a deterministic outcome driven by competitive exclusion by the Cit+ ecotype. The extinction also cannot be explained by demographic stochasticity alone, as the population size of the Cit-ecotype should have been many thousands of cells even during the daily transfer events. Instead, we infer that the extinction must have been caused by a rare chance event in which some aspect of the experimental conditions was inadvertently perturbed.},
keywords = {Citrate Evolution, Demography and Ecology, Historical Contingency},
pubstate = {published},
tppubtype = {article}
}
Quandt E M; Gollihar J; Blount Z D; Ellington A D; Georgiou G; Barrick J E
Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. Journal Article
eLife, 4 (October), pp. e09696, 2015, ISSN: 2050-084X.
Abstract | Links | BibTeX | Altmetric | Tags: Citrate Evolution, Genotypes and Phenotypes, Historical Contingency
@article{Quandt2015,
title = {Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment.},
author = {Erik M. Quandt and Jimmy Gollihar and Zachary D. Blount and Andrew D. Ellington and George Georgiou and Jeffrey E. Barrick},
url = {http://www.ncbi.nlm.nih.gov/pubmed/26465114
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4718724},
doi = {10.7554/eLife.09696},
issn = {2050-084X},
year = {2015},
date = {2015-10-01},
urldate = {2015-10-01},
journal = {eLife},
volume = {4},
number = {October},
pages = {e09696},
abstract = {Evolutionary innovations that enable organisms to colonize new ecological niches are rare compared to gradual evolutionary changes in existing traits. We discovered that key mutations in the \textit{gltA} gene, which encodes citrate synthase (CS), occurred both before and after \textit{Escherichia coli} gained the ability to grow aerobically on citrate (Cit(+) phenotype) during the Lenski long-term evolution experiment. The first \textit{gltA} mutation, which increases CS activity by disrupting NADH-inhibition of this enzyme, is beneficial for growth on the acetate and contributed to preserving the rudimentary Cit(+) trait from extinction when it first evolved. However, after Cit(+) was refined by further mutations, this potentiating \textit{gltA} mutation became deleterious to fitness. A second wave of beneficial \textit{gltA} mutations then evolved that reduced CS activity to below the ancestral level. Thus, dynamic reorganization of central metabolism made colonizing this new nutrient niche contingent on both co-opting and overcoming a history of prior adaptation.},
keywords = {Citrate Evolution, Genotypes and Phenotypes, Historical Contingency},
pubstate = {published},
tppubtype = {article}
}
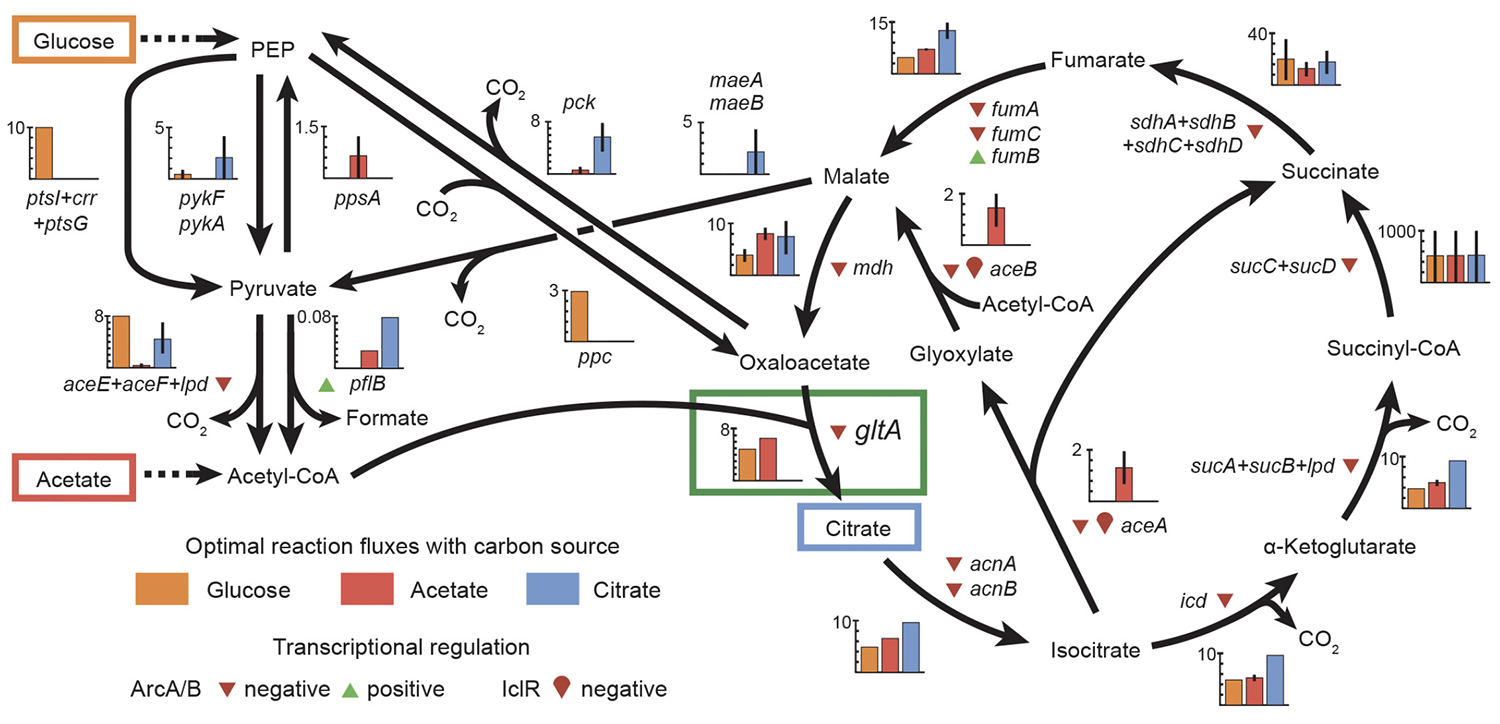
Satterwhite R S; Cooper T F
Constraints on adaptation of Escherichia coli to mixed-resource environments increase over time Journal Article
Evolution, 69 (8), pp. 2067–2078, 2015, ISSN: 00143820.
Abstract | Links | BibTeX | Altmetric | Tags: Descendant Experiments
@article{Satterwhite2015,
title = {Constraints on adaptation of \textit{Escherichia coli} to mixed-resource environments increase over time},
author = {Rebecca S. Satterwhite and Tim F. Cooper},
url = {https://onlinelibrary.wiley.com/doi/10.1111/evo.12710},
doi = {10.1111/evo.12710},
issn = {00143820},
year = {2015},
date = {2015-08-01},
urldate = {2015-08-01},
journal = {Evolution},
volume = {69},
number = {8},
pages = {2067--2078},
abstract = {Can a population evolved in two resources reach the same fitness in both as specialist populations evolved in each of the individual resources? This question is central to theories of ecological specialization, the maintenance of genetic variation, and sympatric speciation, yet relatively few experiments have examined costs of generalism over long-term adaptation. We tested whether selection in environments containing two resources limits a population's ability to adapt to the individual resources by comparing the fitness of replicate \textit{Escherichia coli} populations evolved for 6000 generations in the presence of glucose or lactose alone (specialists), or in varying presentations of glucose and lactose together (generalists). We found that all populations had significant fitness increases in both resources, though the magnitude and rate of these increases differed. For the first 4000 generations, most generalist populations increased in fitness as quickly in the individual resources as the corresponding specialist populations. From 5000 generations, however, a widespread cost of adaptation affected all generalists, indicating a growing constraint on their abilities to adapt to two resources simultaneously. Our results indicate that costs of generalism are prevalent, but may influence evolutionary trajectories only after a period of cost-free adaptation.},
keywords = {Descendant Experiments},
pubstate = {published},
tppubtype = {article}
}
Fox J W; Lenski R E
From Here to Eternity—The Theory and Practice of a Really Long Experiment Journal Article
PLOS Biology, 13 (3), pp. e1002185, 2015, ISSN: 1545-7885.
Abstract | Links | BibTeX | Altmetric | Tags: Review Articles
@article{nokey,
title = {From Here to Eternity—The Theory and Practice of a Really Long Experiment},
author = {Jeremy W. Fox and Richard E. Lenski},
url = {https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002185},
doi = {10.1371/journal.pbio.1002185},
issn = {1545-7885},
year = {2015},
date = {2015-06-23},
urldate = {2015-06-23},
journal = {PLOS Biology},
volume = {13},
number = {3},
pages = { e1002185},
abstract = {In February 1988, Richard Lenski set up 12 replicate populations of a single genotype of \textit{Escherichia coli} in a simple nutrient medium. He has been following their evolution ever since. Here, Lenski answers provocative questions from Jeremy Fox about his iconic "Long-Term Evolution Experiment" (LTEE). The LTEE is a remarkable case study of the interplay of determinism and chance in evolution—and in the conduct of science.},
keywords = {Review Articles},
pubstate = {published},
tppubtype = {article}
}