2024
uz-Zaman Md H; D'Alton S; Barrick J E; Ochman H
Promoter capture drives the emergence of proto-genes in Escherichia coli Journal Article
PLoS Biology, 22 , pp. e3002418, 2024.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution
@article{uz-Zaman2024,
title = {Promoter capture drives the emergence of proto-genes in \textit{Escherichia coli}},
author = {Md. Hassan uz-Zaman and Simon D'Alton and Jeffrey E. Barrick and Howard Ochman},
doi = {10.1371/journal.pbio.3002418},
year = {2024},
date = {2024-05-07},
urldate = {2023-11-17},
journal = {PLoS Biology},
volume = {22},
pages = {e3002418},
publisher = {Cold Spring Harbor Laboratory},
abstract = {The phenomenon of de novo gene birth-the emergence of genes from non-genic sequences-has received considerable attention due to the widespread occurrence of genes that are unique to particular species or genomes. Most instances of de novo gene birth have been recognized through comparative analyses of genome sequences in eukaryotes, despite the abundance of novel, lineage-specific genes in bacteria and the relative ease with which bacteria can be studied in an experimental context. Here, we explore the genetic record of the \textit{Escherichia coli} long-term evolution experiment (LTEE) for changes indicative of "proto-genic" phases of new gene birth in which non-genic sequences evolve stable transcription and/or translation. Over the time span of the LTEE, non-genic regions are frequently transcribed, translated and differentially expressed, with levels of transcription across low-expressed regions increasing in later generations of the experiment. Proto-genes formed downstream of new mutations result either from insertion element activity or chromosomal translocations that fused preexisting regulatory sequences to regions that were not expressed in the LTEE ancestor. Additionally, we identified instances of proto-gene emergence in which a previously unexpressed sequence was transcribed after formation of an upstream promoter, although such cases were rare compared to those caused by recruitment of preexisting promoters. Tracing the origin of the causative mutations, we discovered that most occurred early in the history of the LTEE, often within the first 20,000 generations, and became fixed soon after emergence. Our findings show that proto-genes emerge frequently within evolving populations, can persist stably, and can serve as potential substrates for new gene formation. },
keywords = {Genome Evolution},
pubstate = {published},
tppubtype = {article}
}
2023
Lenski R E
Revisiting the design of the long-term evolution experiment with Escherichia coli Journal Article
Journal of Molecular Evolution, 91 (3), pp. 241-253, 2023.
Abstract | Links | BibTeX | Altmetric | Tags: Citrate Evolution, Descendant Experiments, Fitness Trajectories, Genome Evolution, Methods and Miscellaneous, Review Articles
@article{lenski2023,
title = {Revisiting the design of the long-term evolution experiment with \textit{Escherichia coli}},
author = {Richard E. Lenski},
url = {https://link.springer.com/epdf/10.1007/s00239-023-10095-3?sharing_token=zmDHuK0kbvnJBQq1k96fe_e4RwlQNchNByi7wbcMAY53KNkhv6F2YgRIeC8sZGNejxJrvlAGZWInruED5Dqdai5WeU2RAWL2PJNp0pL9QJO39B_ijCtRZcaW8jqM7PclDJfFwL_78U5zNlQYyCOsQwa1Yxha61uXUWhW-Buiq7o=},
doi = {10.1007/s00239-023-10095-3},
year = {2023},
date = {2023-06-01},
urldate = {2023-02-15},
journal = {Journal of Molecular Evolution},
volume = {91},
number = {3},
pages = {241-253},
abstract = {The long-term evolution experiment (LTEE) with \textit{Escherichia coli} began in 1988 and it continues to this day, with its 12 populations having recently reached 75,000 generations of evolution in a simple, well-controlled environment. The LTEE was designed to explore open-ended questions about the dynamics and repeatability of phenotypic and genetic evolution. Here I discuss various aspects of the LTEE’s experimental design that have enabled its stability and success, including the choices of the culture regime, growth medium, ancestral strain, and statistical replication. I also discuss some of the challenges associated with a long-running project, such as handling procedural errors (e.g., cross-contamination) and managing the expanding collection of frozen samples. The simplicity of the experimental design and procedures have supported the long-term stability of the LTEE. That stability—along with the inherent creativity of the evolutionary process and the emergence of new genomic technologies—provides a platform that has allowed talented students and collaborators to pose questions, collect data, and make discoveries that go far beyond anything I could have imagined at the start of the LTEE.},
keywords = {Citrate Evolution, Descendant Experiments, Fitness Trajectories, Genome Evolution, Methods and Miscellaneous, Review Articles},
pubstate = {published},
tppubtype = {article}
}
Ascensao J A; Wetmore K M; Good B H; Arkin A P; Hallatschek O
Quantifying the adaptive potential of a nascent bacterial community Journal Article
Nature Communications, 14 (1), pp. 248, 2023.
Abstract | Links | BibTeX | Altmetric | Tags: Demography and Ecology, Descendant Experiments, Genome Evolution, Genotypes and Phenotypes
@article{ascensao2023,
title = {Quantifying the adaptive potential of a nascent bacterial community},
author = {Joao A. Ascensao and Kelly M. Wetmore and Benjamin H. Good and Adam P. Arkin and Oskar Hallatschek},
url = {https://www.nature.com/articles/s41467-022-35677-5},
doi = {10.1038/s41467-022-35677-5},
year = {2023},
date = {2023-01-16},
urldate = {2022-01-01},
journal = {Nature Communications},
volume = {14},
number = {1},
pages = {248},
publisher = {Cold Spring Harbor Laboratory},
abstract = {The fitness effects of all possible mutations available to an organism largely shape the dynamics of evolutionary adaptation. Yet, whether and how this adaptive landscape changes over evolutionary times, especially upon ecological diversification and changes in community composition, remains poorly understood. We sought to fill this gap by analyzing a stable community of two closely related ecotypes (“L” and “S”) shortly after they emerged within the \textit{E. coli} Long-Term Evolution Experiment (LTEE). We engineered genome-wide barcoded transposon libraries to measure the invasion fitness effects of all possible gene knockouts in the coexisting strains as well as their ancestor, for many different, ecologically relevant conditions. We find consistent statistical patterns of fitness effect variation across both genetic background and community composition, despite the idiosyncratic behavior of individual knockouts. Additionally, fitness effects are correlated with evolutionary outcomes for a number of conditions, possibly revealing shifting patterns of adaptation. Together, our results reveal how ecological and epistatic effects combine to shape the adaptive landscape in a nascent ecological community.},
keywords = {Demography and Ecology, Descendant Experiments, Genome Evolution, Genotypes and Phenotypes},
pubstate = {published},
tppubtype = {article}
}
2022
Maddamsetti R; Grant N A
Discovery of positive and purifying selection in metagenomic time series of hypermutator microbial populations Journal Article
PLOS Genetics, 18 (8), pp. e1010324, 2022, ISSN: 1553-7404.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution, Mutation Rates
@article{maddamsetti2022,
title = {Discovery of positive and purifying selection in metagenomic time series of hypermutator microbial populations},
author = { Rohan Maddamsetti and Nkrumah A. Grant},
editor = {Jianzhi Zhang},
url = {https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1010324},
doi = {10.1371/journal.pgen.1010324},
issn = {1553-7404},
year = {2022},
date = {2022-08-18},
urldate = {2022-08-18},
journal = {PLOS Genetics},
volume = {18},
number = {8},
pages = {e1010324},
abstract = {A general method to infer both positive and purifying selection during the real-time evolution of hypermutator pathogens would be broadly useful. To this end, we introduce a simple test to infer mode of selection (STIMS) from metagenomic time series of evolving microbial populations. We test STIMS on metagenomic data generated by simulations of bacterial evolution, and on metagenomic data spanning 62,750 generations of Lenski’s long-term evolution experiment with \textit{Escherichia coli} (LTEE). This benchmarking shows that STIMS detects positive selection in both nonmutator and hypermutator populations, and purifying selection in hypermutator populations. Using STIMS, we find strong evidence of ongoing positive selection on key regulators of the \textit{E. coli} gene regulatory network, even in some hypermutator populations. STIMS also detects positive selection on regulatory genes in hypermutator populations of \textit{Pseudomonas aeruginosa} that adapted to subinhibitory concentrations of colistin—an antibiotic of last resort—for just twenty-six days of laboratory evolution. Our results show that the fine-tuning of gene regulatory networks is a general mechanism for rapid and ongoing adaptation. The simplicity of STIMS, together with its intuitive visual interpretation, make it a useful test for positive and purifying selection in metagenomic data sets that track microbial evolution in real-time.},
key = {Maddamsetti2022},
keywords = {Genome Evolution, Mutation Rates},
pubstate = {published},
tppubtype = {article}
}
Jordan J A; Lenski R E; Card K J
Idiosyncratic fitness costs of ampicillin-resistant mutants derived from a long-term experiment with Escherichia coli Journal Article
Antibiotics, 11 (3), pp. 347, 2022, ISSN: 2079-6382.
Abstract | Links | BibTeX | Altmetric | Tags: Descendant Experiments, Genome Evolution, Historical Contingency
@article{Jordan2022.02.06.479266,
title = {Idiosyncratic fitness costs of ampicillin-resistant mutants derived from a long-term experiment with \textit{Escherichia coli}},
author = {Jalin A. Jordan and Richard E. Lenski and Kyle J. Card},
url = {https://www.biorxiv.org/content/10.1101/2022.02.06.479266v1},
doi = {10.3390/antibiotics11030347},
issn = {2079-6382},
year = {2022},
date = {2022-03-13},
urldate = {2022-03-13},
journal = {Antibiotics},
volume = {11},
number = {3},
pages = {347},
publisher = {Cold Spring Harbor Laboratory},
abstract = {Antibiotic resistance is a growing concern that has prompted a renewed focus on drug discovery, stewardship, and evolutionary studies of the patterns and processes that underlie this phenomenon. A resistant strain’s competitive fitness relative to its sensitive counterparts in the absence of drug can impact its spread and persistence in both clinical and community settings. In a prior study, we examined the fitness of tetracycline-resistant clones that evolved from five different Escherichia coli genotypes, which had diverged during a long-term evolution experiment. In this study, we build on that work to examine whether ampicillin-resistant mutants are also less fit in the absence of the drug than their sensitive parents, and whether the cost of resistance is constant or variable among independently derived lines. Like the tetracycline-resistant lines, the ampicillin-resistant mutants were often less fit than their sensitive parents, with significant variation in the fitness costs among the mutants. This variation was not associated with the level of resistance conferred by the mutations, nor did it vary across the different parental backgrounds. In our earlier study, some of the variation in fitness costs associated with tetracycline resistance was explained by the effects of different mutations affecting the same cellular pathway and even the same gene. In contrast, the variance among the ampicillin-resistant mutants was associated with different sets of target genes. About half of the resistant clones suffered large fitness deficits, and their mutations impacted major outer-membrane proteins or subunits of RNA polymerases. The other mutants experienced little or no fitness costs and with, one exception, they had mutations affecting other genes and functions. Our findings underscore the importance of comparative studies on the evolution of antibiotic resistance, and they highlight the nuanced processes that shape these phenotypes.},
keywords = {Descendant Experiments, Genome Evolution, Historical Contingency},
pubstate = {published},
tppubtype = {article}
}
2021
Consuegra J; Gaffé J; Lenski R E; Hindré T; Barrick J E; Tenaillon O; Schneider D
Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria Journal Article
Nature Communications, 12 (1), pp. 980, 2021, ISSN: 2041-1723.
Abstract | Links | BibTeX | Altmetric | Tags: Fitness Trajectories, Genome Evolution, Mutation Rates
@article{Consuegra2021,
title = {Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria},
author = {Jessika Consuegra and Joël Gaffé and Richard E. Lenski and Thomas Hindré and Jeffrey E. Barrick and Olivier Tenaillon and Dominique Schneider},
url = {http://www.nature.com/articles/s41467-021-21210-7},
doi = {https://doi.org/10.1038/s41467-021-21210-7},
issn = {2041-1723},
year = {2021},
date = {2021-12-01},
urldate = {2021-12-01},
journal = {Nature Communications},
volume = {12},
number = {1},
pages = {980},
publisher = {Springer US},
abstract = {Insertion sequences (IS) are ubiquitous bacterial mobile genetic elements, and the mutations they cause can be deleterious, neutral, or beneficial. The long-term dynamics of IS elements and their effects on bacteria are poorly understood, including whether they are primarily genomic parasites or important drivers of adaptation by natural selection. Here, we investigate the dynamics of IS elements and their contribution to genomic evolution and fitness during a long-term experiment with \textit{Escherichia coli}. IS elements account for ~35% of the mutations that reached high frequency through 50,000 generations in those populations that retained the ancestral point-mutation rate. In mutator populations, IS-mediated mutations are only half as frequent in absolute numbers. In one population, an exceptionally high ~8-fold increase in IS 150 copy number is associated with the beneficial effects of early insertion mutations; however, this expansion later slowed down owing to reduced IS 150 activity. This population also achieves the lowest fitness, suggesting that some avenues for further adaptation are precluded by the IS 150 -mediated mutations. More generally, across all populations, we find that higher IS activity becomes detrimental to adaptation over evolutionary time. Therefore, IS-mediated mutations can both promote and constrain evolvability.},
keywords = {Fitness Trajectories, Genome Evolution, Mutation Rates},
pubstate = {published},
tppubtype = {article}
}
Grant N A; Maddamsetti R; Lenski R E
Maintenance of Metabolic Plasticity despite Relaxed Selection in a Long-Term Evolution Experiment with Escherichia coli Journal Article
The American Naturalist, 198 (1), pp. 93–112, 2021, ISSN: 0003-0147.
Abstract | Links | BibTeX | Altmetric | Tags: Correlated Responses, Fitness Trajectories, Genome Evolution, Genotypes and Phenotypes
@article{Grant2021b,
title = {Maintenance of Metabolic Plasticity despite Relaxed Selection in a Long-Term Evolution Experiment with \textit{Escherichia coli}},
author = {Nkrumah A. Grant and Rohan Maddamsetti and Richard E. Lenski},
url = {https://www.journals.uchicago.edu/doi/10.1086/714530},
doi = {10.1086/714530},
issn = {0003-0147},
year = {2021},
date = {2021-07-01},
urldate = {2021-07-01},
journal = {The American Naturalist},
volume = {198},
number = {1},
pages = {93--112},
abstract = {Traits that are unused in a given environment are subject to processes that tend to erode them, leading to reduced fitness in other environments. Although this general tendency is clear, we know much less about why some traits are lost while others are retained and about the roles of mutation and selection in generating different responses. We addressed these issues by examining populations of a facultative anaerobe, \textit{Escherichia coli}, that have evolved for 130 years in the presence of oxygen, with relaxed selection for anaerobic growth and the associated metabolic plasticity. We asked whether evolution led to the loss, improvement, or maintenance of anaerobic growth, and we analyzed gene expression and mutational data sets to understand the outcomes. We identified genomic signatures of both positive and purifying selection on aerobic-specific genes, while anaerobic-specific genes showed clear evidence of relaxed selection. We also found parallel evolution at two interacting loci that regulate anaerobic growth. We competed the ancestor and evolved clones from each population in an anoxic environment, and we found that anaerobic fitness had not decayed, despite relaxed selection. In summary, relaxed selection does not necessarily reduce an organism's fitness in other environments. Instead, the genetic architecture of the traits under relaxed selection and their correlations with traits under positive and purifying selection may sometimes determine evolutionary outcomes.},
keywords = {Correlated Responses, Fitness Trajectories, Genome Evolution, Genotypes and Phenotypes},
pubstate = {published},
tppubtype = {article}
}
Card K J; Thomas M D; Graves J L; Barrick J E; Lenski R E
Genomic evolution of antibiotic resistance is contingent on genetic background following a long-term experiment with Escherichia coli Journal Article
Proceedings of the National Academy of Sciences of the United States of America, 118 (5), pp. e2016886118, 2021, ISSN: 0027-8424.
Abstract | Links | BibTeX | Altmetric | Tags: Descendant Experiments, Genome Evolution, Historical Contingency
@article{Card2021,
title = {Genomic evolution of antibiotic resistance is contingent on genetic background following a long-term experiment with \textit{Escherichia coli}},
author = {Kyle J. Card and Misty D. Thomas and Joseph L. Graves and Jeffrey E. Barrick and Richard E. Lenski},
url = {http://www.pnas.org/lookup/doi/10.1073/pnas.2016886118},
doi = {10.1073/pnas.2016886118},
issn = {0027-8424},
year = {2021},
date = {2021-02-01},
urldate = {2021-02-01},
journal = {Proceedings of the National Academy of Sciences of the United States of America},
volume = {118},
number = {5},
pages = {e2016886118},
abstract = {Antibiotic resistance is a growing health concern. Efforts to control resistance would benefit from an improved ability to forecast when and how it will evolve. Epistatic interactions between mutations can promote divergent evolutionary trajectories, which complicates our ability to predict evolution. We recently showed that differences between genetic backgrounds can lead to idiosyncratic responses in the evolvability of phenotypic resistance, even among closely related \textit{Escherichia coli} strains. In this study, we examined whether a strain's genetic background also influences the genotypic evolution of resistance. Do lineages founded by different genotypes take parallel or divergent mutational paths to achieve their evolved resistance states? We addressed this question by sequencing the complete genomes of antibiotic-resistant clones that evolved from several different genetic starting points during our earlier experiments. We first validated our statistical approach by quantifying the specificity of genomic evolution with respect to antibiotic treatment. As expected, mutations in particular genes were strongly associated with each drug. Then, we determined that replicate lines evolved from the same founding genotypes had more parallel mutations at the gene level than lines evolved from different founding genotypes, although these effects were more subtle than those showing antibiotic specificity. Taken together with our previous work, we conclude that historical contingency can alter both genotypic and phenotypic pathways to antibiotic resistance.},
keywords = {Descendant Experiments, Genome Evolution, Historical Contingency},
pubstate = {published},
tppubtype = {article}
}
Maddamsetti R
Selection Maintains Protein Interactome Resilience in the Long-Term Evolution Experiment with Escherichia coli Journal Article
Genome Biology and Evolution, 13 (6), pp. evab074, 2021.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution, Genotypes and Phenotypes
@article{Maddamsetti2021,
title = {Selection Maintains Protein Interactome Resilience in the Long-Term Evolution Experiment with \emph{Escherichia coli}},
author = {Rohan Maddamsetti},
url = {https://academic.oup.com/gbe/article/13/6/evab074/6240992},
doi = {https://doi.org/10.1093/gbe/evab074},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Genome Biology and Evolution},
volume = {13},
number = {6},
pages = {evab074},
abstract = {Most cellular functions are carried out by a dynamic network of interacting proteins. An open question is whether the network properties of protein interactomes represent phenotypes under natural selection. One proposal is that protein interactomes have evolved to be resilient, such that they tend to maintain connectivity when proteins are removed from the network. This hypothesis predicts that interactome resilience should be maintained by natural selection during long-term experimental evolution. I tested this prediction by modeling the evolution of protein–protein interaction (PPI) networks in Lenski’s long-term evolution experiment with \textit{Escherichia coli} (LTEE). In this test, I removed proteins affected by nonsense, insertion, deletion, and transposon mutations in evolved LTEE strains, and measured the resilience of the resulting networks. I compared the rate of change of network resilience in each LTEE population to the rate of change of network resilience for corresponding randomized networks. The evolved PPI networks are significantly more resilient than networks in which random proteins have been deleted. Moreover, the evolved networks are generally more resilient than networks in which the random deletion of proteins was restricted to those disrupted in LTEE. These results suggest that evolution in the LTEE has favored PPI networks that are, on average, more resilient than expected from the genetic variation across the evolved strains. My findings therefore support the hypothesis that selection maintains protein interactome resilience over evolutionary time.},
keywords = {Genome Evolution, Genotypes and Phenotypes},
pubstate = {published},
tppubtype = {article}
}
Maddamsetti R
Universal Constraints on Protein Evolution in the Long-Term Evolution Experiment with Escherichia coli Journal Article
Genome Biology and Evolution, 13 (6), pp. evab070, 2021.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution, Genotypes and Phenotypes
@article{Maddamsetti2021b,
title = {Universal Constraints on Protein Evolution in the Long-Term Evolution Experiment with \textit{Escherichia coli}},
author = {Rohan Maddamsetti},
url = {https://academic.oup.com/gbe/article/13/6/evab070/6226398},
doi = {https://doi.org/10.1093/gbe/evab070},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Genome Biology and Evolution},
volume = {13},
number = {6},
pages = {evab070},
abstract = {Although it is well known that abundant proteins evolve slowly across the tree of life, there is little consensus for why this is true. Here, I report that abundant proteins evolve slowly in the hypermutator populations of Lenski’s long-term evolution experiment with \textit{Escherichia coli} (LTEE). Specifically, the density of all observed mutations per gene, as measured in metagenomic time series covering 60,000 generations of the LTEE, significantly anticorrelates with mRNA abundance, protein abundance, and degree of protein–protein interaction. The same pattern holds for nonsynonymous mutation density. However, synonymous mutation density, measured across the LTEE hypermutator populations, positively correlates with protein abundance. These results show that universal constraints on protein evolution are visible in data spanning three decades of experimental evolution. Therefore, it should be possible to design experiments to answer why abundant proteins evolve slowly.},
keywords = {Genome Evolution, Genotypes and Phenotypes},
pubstate = {published},
tppubtype = {article}
}
2020
Maddamsetti R; Grant N A
Divergent Evolution of Mutation Rates and Biases in the Long-Term Evolution Experiment with Escherichia coli Journal Article
Genome Biology and Evolution, 12 (9), pp. 1591-1603, 2020, ISSN: 1759-6653.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution, Mutation Rates
@article{nokey,
title = {Divergent Evolution of Mutation Rates and Biases in the Long-Term Evolution Experiment with \textit{Escherichia coli}},
author = {Rohan Maddamsetti and Nkrumah A. Grant},
editor = {George Zhang},
url = {https://academic.oup.com/gbe/article/12/9/1591/5898197},
doi = {10.1093/gbe/evaa178},
issn = {1759-6653},
year = {2020},
date = {2020-08-21},
urldate = {2020-08-21},
journal = {Genome Biology and Evolution},
volume = {12},
number = {9},
pages = {1591-1603},
abstract = {All organisms encode enzymes that replicate, maintain, pack, recombine, and repair their genetic material. For this reason, mutation rates and biases also evolve by mutation, variation, and natural selection. By examining metagenomic time series of the Lenski long-term evolution experiment (LTEE) with \textit{Escherichia coli} (Good BH, McDonald MJ, Barrick JE, Lenski RE, Desai MM. 2017. The dynamics of molecular evolution over 60,000 generations. Nature 551(7678):45–50.), we find that local mutation rate variation has evolved during the LTEE. Each LTEE population has evolved idiosyncratic differences in their rates of point mutations, indels, and mobile element insertions, due to the fixation of various hypermutator and antimutator alleles. One LTEE population, called Ara+3, shows a strong, symmetric wave pattern in its density of point mutations, radiating from the origin of replication. This pattern is largely missing from the other LTEE populations, most of which evolved missense, indel, or structural mutations in \textit{topA}, \textit{fis}, and \textit{dusB}—loci that all affect DNA topology. The distribution of mutations in those genes over time suggests epistasis and historical contingency in the evolution of DNA topology, which may have in turn affected local mutation rates. Overall, the replicate populations of the LTEE have largely diverged in their mutation rates and biases, even though they have adapted to identical abiotic conditions.},
keywords = {Genome Evolution, Mutation Rates},
pubstate = {published},
tppubtype = {article}
}
2018
Maddamsetti R; Lenski R E
PLOS Genetics, 14 (1), pp. e1007199, 2018, ISSN: 1553-7404.
Abstract | Links | BibTeX | Altmetric | Tags: Descendant Experiments, Genome Evolution
@article{Maddamsetti2018,
title = {Analysis of bacterial genomes from an evolution experiment with horizontal gene transfer shows that recombination can sometimes overwhelm selection},
author = {Rohan Maddamsetti and Richard E. Lenski},
editor = {Ivan Matic},
url = {https://dx.plos.org/10.1371/journal.pgen.1007199},
doi = {10.1371/journal.pgen.1007199},
issn = {1553-7404},
year = {2018},
date = {2018-01-01},
urldate = {2018-01-01},
journal = {PLOS Genetics},
volume = {14},
number = {1},
pages = {e1007199},
abstract = {Few experimental studies have examined the role that sexual recombination plays in bacterial evolution, including the effects of horizontal gene transfer on genome structure. To address this limitation, we analyzed genomes from an experiment in which \textit{Escherichia coli} K-12 Hfr (high frequency recombination) donors were periodically introduced into 12 evolving populations of \textit{E. coli} B and allowed to conjugate repeatedly over the course of 1000 generations. Previous analyses of the evolved strains from this experiment showed that recombination did not accelerate adaptation, despite increasing genetic variation relative to asexual controls. However, the resolution in that previous work was limited to only a few genetic markers. We sought to clarify and understand these puzzling results by sequencing complete genomes from each population. The effects of recombination were highly variable: one lineage was mostly derived from the donors, while another acquired almost no donor DNA. In most lineages, some regions showed repeated introgression and others almost none. Regions with high introgression tended to be near the donors' origin of transfer sites. To determine whether introgressed alleles imposed a genetic load, we extended the experiment for 200 generations without recombination and sequenced whole-population samples. Beneficial alleles in the recipient populations were occasionally driven extinct by maladaptive donor-derived alleles. On balance, our analyses indicate that the plasmid-mediated recombination was sufficiently frequent to drive donor alleles to fixation without providing much, if any, selective advantage.},
keywords = {Descendant Experiments, Genome Evolution},
pubstate = {published},
tppubtype = {article}
}
2017
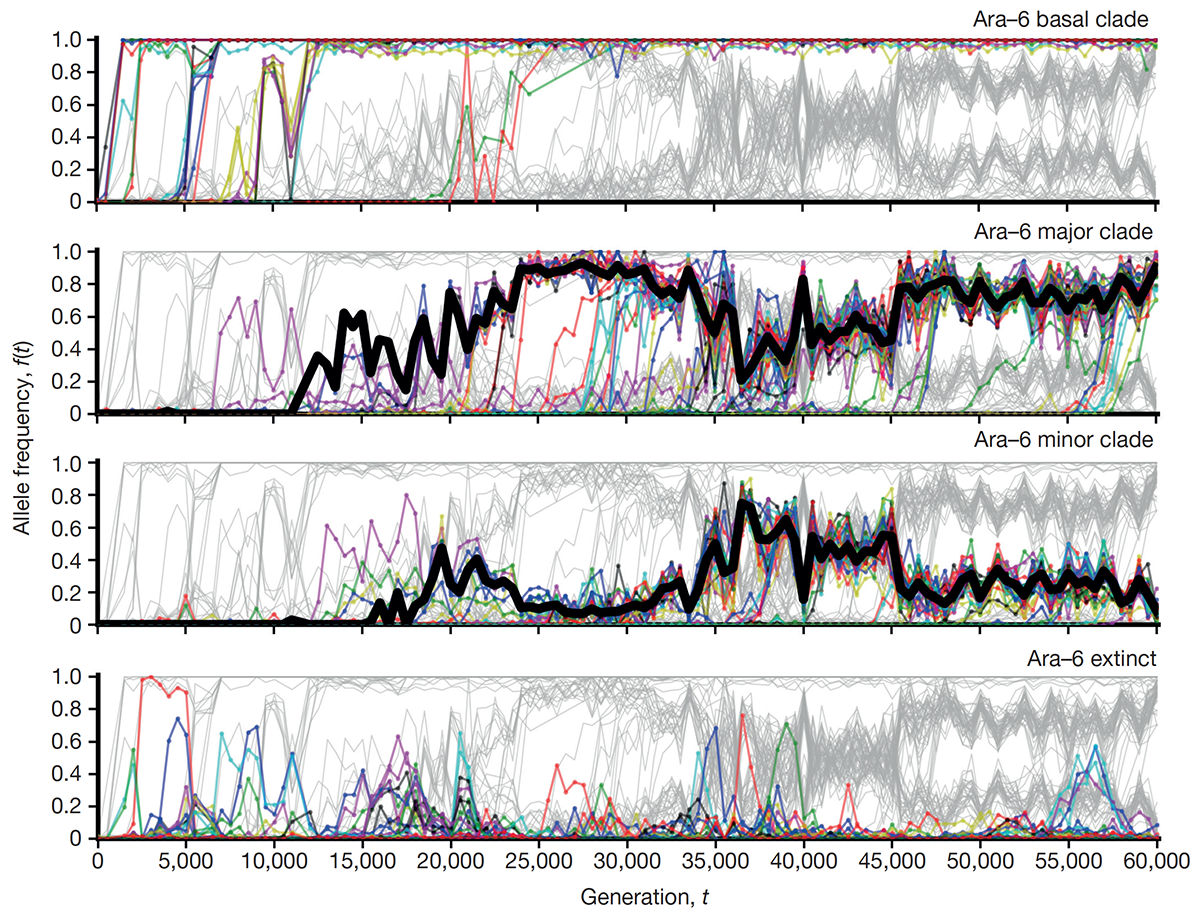
Good B H; McDonald M J; Barrick J E; Lenski R E; Desai M M
The dynamics of molecular evolution over 60,000 generations Journal Article
Nature, 551 (7678), pp. 45–50, 2017, ISSN: 14764687.
Abstract | Links | BibTeX | Altmetric | Tags: Demography and Ecology, Genome Evolution, Historical Contingency, Mutation Rates, Parallelism and Divergence
@article{Good2017,
title = {The dynamics of molecular evolution over 60,000 generations},
author = {Benjamin H. Good and Michael J. McDonald and Jeffrey E. Barrick and Richard E. Lenski and Michael M. Desai},
url = {http://dx.doi.org/10.1038/nature24287},
doi = {10.1038/nature24287},
issn = {14764687},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {Nature},
volume = {551},
number = {7678},
pages = {45--50},
publisher = {Nature Publishing Group},
abstract = {The outcomes of evolution are determined by a stochastic dynamical process that governs how mutations arise and spread through a population. However, it is difficult to observe these dynamics directly over long periods and across entire genomes. Here we analyse the dynamics of molecular evolution in twelve experimental populations of \textit{Escherichia coli}, using whole-genome metagenomic sequencing at five hundred-generation intervals through sixty thousand generations. Although the rate of fitness gain declines over time, molecular evolution is characterized by signatures of rapid adaptation throughout the duration of the experiment, with multiple beneficial variants simultaneously competing for dominance in each population. Interactions between ecological and evolutionary processes play an important role, as long-term quasi-stable coexistence arises spontaneously in most populations, and evolution continues within each clade. We also present evidence that the targets of natural selection change over time, as epistasis and historical contingency alter the strength of selection on different genes. Together, these results show that long-term adaptation to a constant environment can be a more complex and dynamic process than is often assumed.},
keywords = {Demography and Ecology, Genome Evolution, Historical Contingency, Mutation Rates, Parallelism and Divergence},
pubstate = {published},
tppubtype = {article}
}
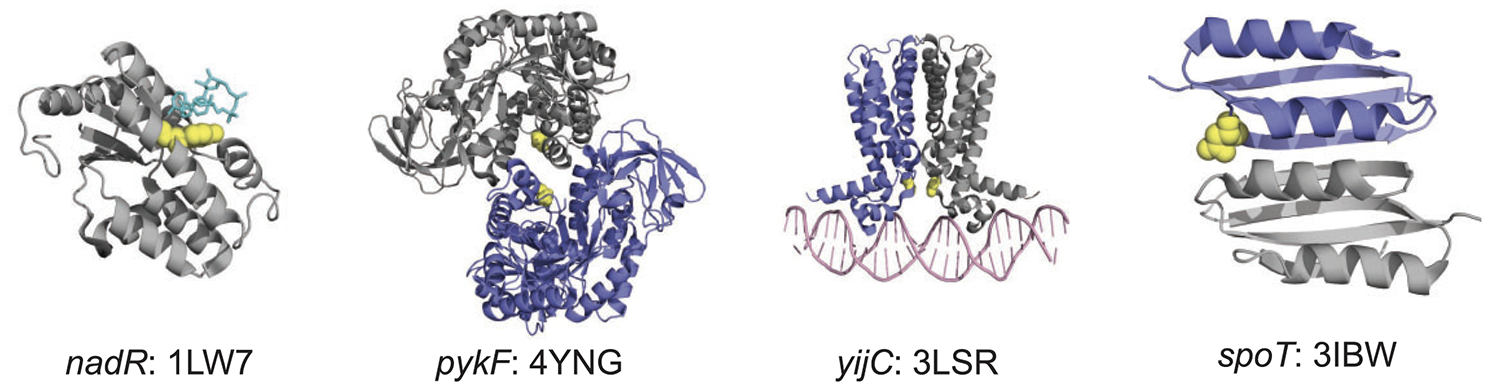
Maddamsetti R; Hatcher P J; Green A G; Williams B L; Marks D S; Lenski R E
Core genes evolve rapidly in the long-term evolution experiment with Escherichia coli Journal Article
Genome Biology and Evolution, 9 (4), pp. 1072–1083, 2017, ISSN: 17596653.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution, Genotypes and Phenotypes, Mutation Rates
@article{Maddamsetti2017,
title = {Core genes evolve rapidly in the long-term evolution experiment with \textit{Escherichia coli}},
author = {Rohan Maddamsetti and Philip J. Hatcher and Anna G. Green and Barry L. Williams and Debora S. Marks and Richard E. Lenski},
url = {https://academic.oup.com/gbe/article/9/4/1072/3100447},
doi = {10.1093/gbe/evx064},
issn = {17596653},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
journal = {Genome Biology and Evolution},
volume = {9},
number = {4},
pages = {1072--1083},
abstract = {Bacteria can evolve rapidly under positive selection owing to their vast numbers, allowing their genes to diversify by adapting to different environments. We asked whether the same genes that evolve rapidly in the long-term evolution experiment (LTEE) with \textit{Escherichia coli} have also diversified extensively in nature. To make this comparison, we identified ~2000 core genes shared among 60 \textit{E. coli} strains. During the LTEE, core genes accumulated significantly more nonsynonymous mutations than flexible (i.e., noncore) genes. Furthermore, core genes under positive selection in the LTEE are more conserved in nature than the average core gene. In some cases, adaptive mutations appear to modify protein functions, rather than merely knocking them out. The LTEE conditions are novel for \textit{E. coli}, at least in relation to its evolutionary history in nature. The constancy and simplicity of the environment likely favor the complete loss of some unused functions and the fine-tuning of others.},
keywords = {Genome Evolution, Genotypes and Phenotypes, Mutation Rates},
pubstate = {published},
tppubtype = {article}
}
2016
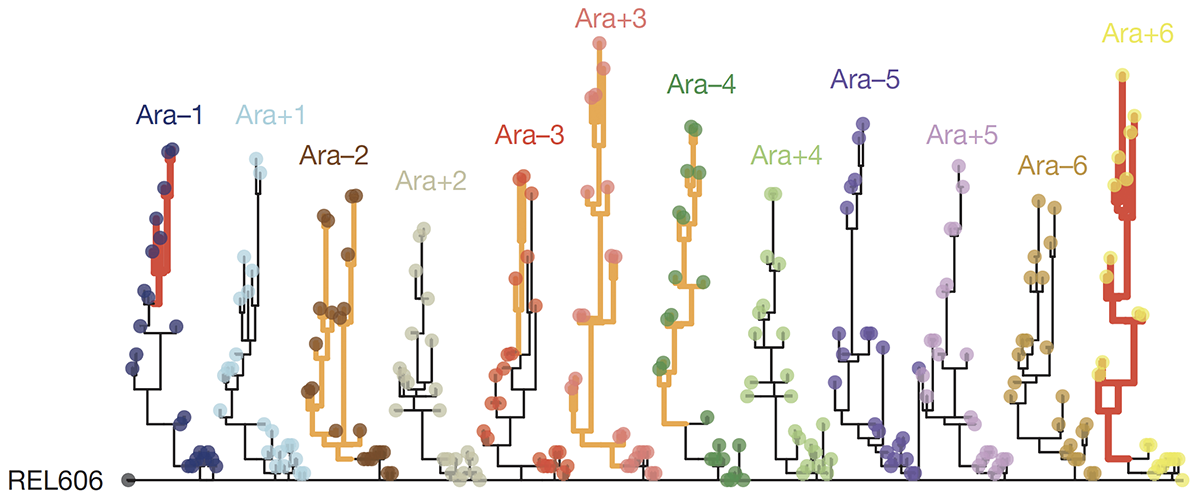
Tenaillon O; Barrick J E; Ribeck N; Deatherage D E; Blanchard J L; Dasgupta A; Wu G C; Wielgoss S; Cruveiller S; Medigue C; Schneider D; Lenski R E
Tempo and mode of genome evolution in a 50,000-generation experiment. Journal Article
Nature, 536 (7615), pp. 165–170, 2016, ISSN: 1476-4687.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution, Mutation Rates, Parallelism and Divergence
@article{Tenaillon2016,
title = {Tempo and mode of genome evolution in a 50,000-generation experiment.},
author = {Olivier Tenaillon and Jeffrey E. Barrick and Noah Ribeck and Daniel E. Deatherage and Jeffrey L. Blanchard and Aurko Dasgupta and Gabriel C. Wu and Sebastien Wielgoss and Stephane Cruveiller and Claudine Medigue and Dominique Schneider and Richard E. Lenski},
url = {http://www.ncbi.nlm.nih.gov/pubmed/27479321},
doi = {10.1038/nature18959},
issn = {1476-4687},
year = {2016},
date = {2016-08-01},
urldate = {2016-08-01},
journal = {Nature},
volume = {536},
number = {7615},
pages = {165--170},
publisher = {Nature Publishing Group},
abstract = {Adaptation by natural selection depends on the rates, effects and interactions of many mutations, making it difficult to determine what proportion of mutations in an evolving lineage are beneficial. Here we analysed 264 complete genomes from 12 \textit{Escherichia coli} populations to characterize their dynamics over 50,000 generations. The populations that retained the ancestral mutation rate support a model in which most fixed mutations are beneficial, the fraction of beneficial mutations declines as fitness rises, and neutral mutations accumulate at a constant rate. We also compared these populations to mutation-accumulation lines evolved under a bottlenecking regime that minimizes selection. Nonsynonymous mutations, intergenic mutations, insertions and deletions are overrepresented in the long-term populations, further supporting the inference that most mutations that reached high frequency were favoured by selection. These results illuminate the shifting balance of forces that govern genome evolution in populations adapting to a new environment.},
keywords = {Genome Evolution, Mutation Rates, Parallelism and Divergence},
pubstate = {published},
tppubtype = {article}
}
2015
Maddamsetti R; Hatcher P J; Cruveiller S; Medigue C; Barrick J E; Lenski R E
Molecular Biology and Evolution, 32 (11), 2015, ISSN: 0737-4038.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution
@article{Maddamsetti2015,
title = {Synonymous Genetic Variation in Natural Isolates of \textit{Escherichia coli} Does Not Predict Where Synonymous Substitutions Occur in a Long-Term Experiment},
author = {Rohan Maddamsetti and Philip J. Hatcher and Stephane Cruveiller and Claudine Medigue and Jeffrey E. Barrick and Richard E. Lenski},
url = {https://academic.oup.com/mbe/article-lookup/doi/10.1093/molbev/msv161},
doi = {10.1093/molbev/msv161},
issn = {0737-4038},
year = {2015},
date = {2015-11-01},
urldate = {2015-11-01},
journal = {Molecular Biology and Evolution},
volume = {32},
number = {11},
abstract = {Synonymous genetic differences vary by more than 20-fold among genes in natural isolates of \textit{Escherichia coli}. One hypothesis to explain this heterogeneity is that genes with high levels of synonymous variation mutate at higher rates than genes with low synonymous variation. If so, then one would expect to observe similar mutational patterns in evolution experiments. In fact, however, the pattern of synonymous substitutions in a long-term evolution experiment with \textit{E. coli} does not support this hypothesis. In particular, the extent of synonymous variation across genes in that experiment does not reflect the variation observed in natural isolates of \textit{E. coli}. Instead, gene length alone predicts with high accuracy the prevalence of synonymous changes in the experimental populations. We hypothesize that patterns of synonymous variation in natural \textit{E. coli} populations are instead caused by differences across genomic regions in their effective population size that, in turn, reflect different histories of recombination, horizontal gene transfer, selection, and population structure.},
keywords = {Genome Evolution},
pubstate = {published},
tppubtype = {article}
}
Maddamsetti R; Lenski R E; Barrick J E
Adaptation, Clonal Interference, and Frequency-Dependent Interactions in a Long-Term Evolution Experiment with Escherichia coli Journal Article
Genetics, 200 (2), pp. 619-631, 2015, ISSN: 1943-2631.
Abstract | Links | BibTeX | Altmetric | Tags: Demography and Ecology, Fitness Trajectories, Genome Evolution
@article{nokey,
title = {Adaptation, Clonal Interference, and Frequency-Dependent Interactions in a Long-Term Evolution Experiment with \emph{Escherichia coli}},
author = {Rohan Maddamsetti and Richard E. Lenski and Jeffrey E. Barrick},
url = {https://academic.oup.com/genetics/article/200/2/619/5936186},
doi = {10.1534/genetics.115.176677},
issn = {1943-2631},
year = {2015},
date = {2015-04-24},
urldate = {2015-04-24},
journal = {Genetics},
volume = {200},
number = {2},
pages = {619-631},
abstract = {Twelve replicate populations of \textit{Escherichia coli} have been evolving in the laboratory for >25 years and 60,000 generations. We analyzed bacteria from whole-population samples frozen every 500 generations through 20,000 generations for one well-studied population, called Ara−1. By tracking 42 known mutations in these samples, we reconstructed the history of this population’s genotypic evolution over this period. The evolutionary dynamics of Ara−1 show strong evidence of selective sweeps as well as clonal interference between competing lineages bearing different beneficial mutations. In some cases, sets of several mutations approached fixation simultaneously, often conveying no information about their order of origination; we present several possible explanations for the existence of these mutational cohorts. Against a backdrop of rapid selective sweeps both earlier and later, two genetically diverged clades coexisted for >6000 generations before one went extinct. In that time, many additional mutations arose in the clade that eventually prevailed. We show that the clades evolved a frequency-dependent interaction, which prevented the immediate competitive exclusion of either clade, but which collapsed as beneficial mutations accumulated in the clade that prevailed. Clonal interference and frequency dependence can occur even in the simplest microbial populations. Furthermore, frequency dependence may generate dynamics that extend the period of coexistence that would otherwise be sustained by clonal interference alone.},
keywords = {Demography and Ecology, Fitness Trajectories, Genome Evolution},
pubstate = {published},
tppubtype = {article}
}
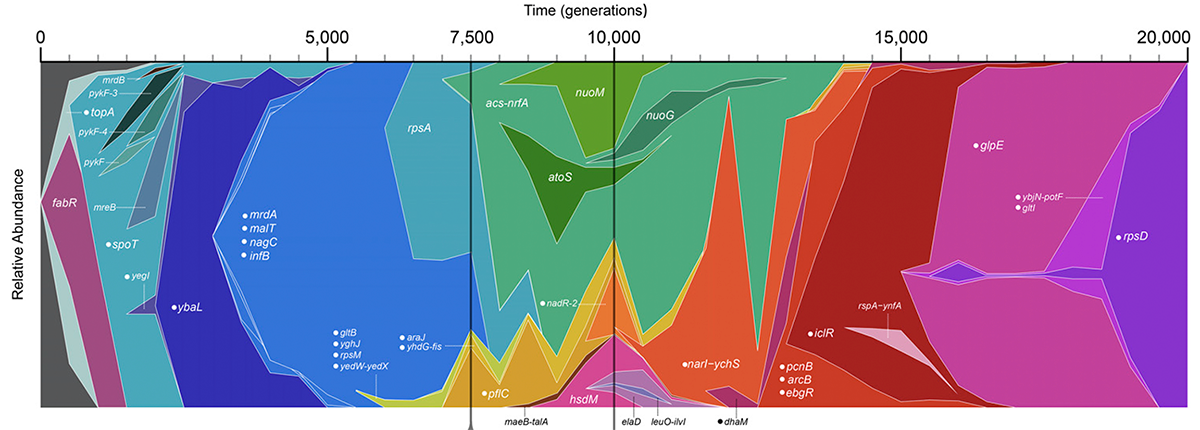
2014
Raeside C; Gaffé J; Deatherage D E; Tenaillon O; Briska A M; Ptashkin R N; Cruveiller S; Médigue C; Lenski R E; Barrick J E; Schneider D
Large Chromosomal Rearrangements during a Long-Term Evolution Experiment with Escherichia coli Journal Article
mBio, 5 (5), pp. 1–13, 2014, ISSN: 2161-2129.
Abstract | Links | BibTeX | Altmetric | Tags: Genome Evolution
@article{Raeside2014,
title = {Large Chromosomal Rearrangements during a Long-Term Evolution Experiment with \textit{Escherichia coli}},
author = {Colin Raeside and Joël Gaffé and Daniel E. Deatherage and Olivier Tenaillon and Adam M. Briska and Ryan N. Ptashkin and Stéphane Cruveiller and Claudine Médigue and Richard E. Lenski and Jeffrey E. Barrick and Dominique Schneider},
editor = {Søren Molin and Fernando Baquero},
url = {https://journals.asm.org/doi/10.1128/mBio.01377-14},
doi = {10.1128/mBio.01377-14},
issn = {2161-2129},
year = {2014},
date = {2014-10-01},
urldate = {2014-10-01},
journal = {mBio},
volume = {5},
number = {5},
pages = {1--13},
abstract = {Large-scale rearrangements may be important in evolution because they can alter chromosome organization and gene expression in ways not possible through point mutations. In a long-term evolution experiment, twelve \textit{Escherichia coli} populations have been propagated in a glucose-limited environment for over 25 years. We used whole-genome mapping (optical mapping) combined with genome sequencing and PCR analysis to identify the large-scale chromosomal rearrangements in clones from each population after 40,000 generations. A total of 110 rearrangement events were detected, including 82 deletions, 19 inversions, and 9 duplications, with lineages having between 5 and 20 events. In three populations, successive rearrangements impacted particular regions. In five populations, rearrangements affected over a third of the chromosome. Most rearrangements involved recombination between insertion sequence (IS) elements, illustrating their importance in mediating genome plasticity. Two lines of evidence suggest that at least some of these rearrangements conferred higher fitness. First, parallel changes were observed across the independent populations, with ~65% of the rearrangements affecting the same loci in at least two populations. For example, the ribose-utilization operon and the \textit{manB} - \textit{cpsG} region were deleted in 12 and 10 populations, respectively, suggesting positive selection, and this inference was previously confirmed for the former case. Second, optical maps from clones sampled over time from one population showed that most rearrangements occurred early in the experiment, when fitness was increasing most rapidly. However, some rearrangements likely occur at high frequency and may have simply hitchhiked to fixation. In any case, large-scale rearrangements clearly influenced genomic evolution in these populations.},
keywords = {Genome Evolution},
pubstate = {published},
tppubtype = {article}
}
2013
Wielgoss S; Barrick J E; Tenaillon O; Wiser M J; Dittmar W J; Cruveiller S; Chane-Woon-Ming B; Médigue C; Lenski R E; Schneider D
Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load Journal Article
Proceedings of the National Academy of Sciences of the United States of America, 110 (1), pp. 222–227, 2013, ISSN: 0027-8424.
Abstract | Links | BibTeX | Altmetric | Tags: Fitness Trajectories, Genome Evolution, Genotypes and Phenotypes, Mutation Rates, Theory and Simulations
@article{Wielgoss2013,
title = {Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load},
author = {Sébastien Wielgoss and Jeffrey E. Barrick and Olivier Tenaillon and Michael J. Wiser and W. James Dittmar and Stéphane Cruveiller and Béatrice Chane-Woon-Ming and Claudine Médigue and Richard E. Lenski and Dominique Schneider},
url = {http://www.pnas.org/cgi/doi/10.1073/pnas.1219574110},
doi = {10.1073/pnas.1219574110},
issn = {0027-8424},
year = {2013},
date = {2013-01-01},
urldate = {2013-01-01},
journal = {Proceedings of the National Academy of Sciences of the United States of America},
volume = {110},
number = {1},
pages = {222--227},
abstract = {Mutations are the ultimate source of heritable variation for evolution. Understanding how mutation rates themselves evolve is thus essential for quantitatively understanding many evolutionary processes. According to theory, mutation rates should be minimized for well-adapted populations living in stable environments, whereas hypermutators may evolve if conditions change. However, the long-term fate of hypermutators is unknown. Using a phylogenomic approach, we found that an adapting \textit{Escherichia coli} population that first evolved a \textit{mutT} hypermutator phenotype was later invaded by two independent lineages with \textit{mutY} mutations that reduced genome-wide mutation rates. Applying neutral theory to synonymous substitutions, we dated the emergence of these mutations and inferred that the \textit{mutT} mutation increased the point-mutation rate by ~150-fold, whereas the \textit{mutY} mutations reduced the rate by ~40-60%, with a corresponding decrease in the genetic load. Thus, the long-term fate of the hypermutators was governed by the selective advantage arising from a reduced mutation rate as the potential for further adaptation declined.},
keywords = {Fitness Trajectories, Genome Evolution, Genotypes and Phenotypes, Mutation Rates, Theory and Simulations},
pubstate = {published},
tppubtype = {article}
}
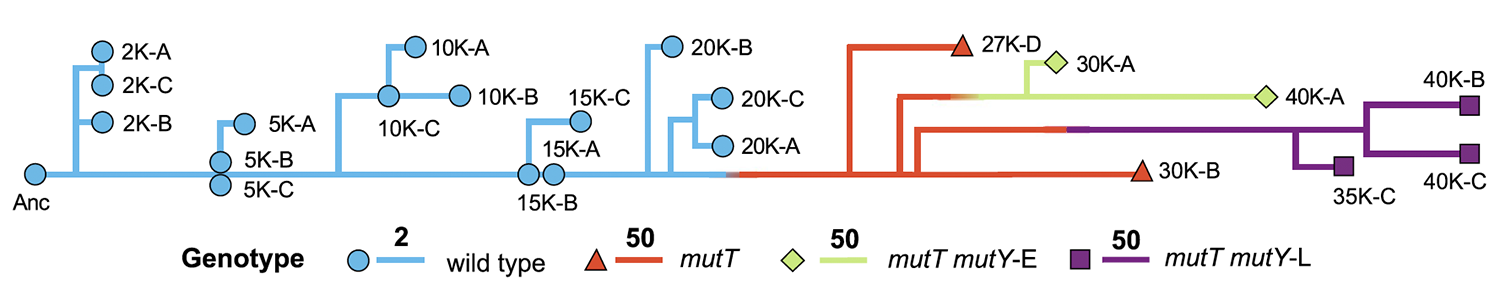
2012
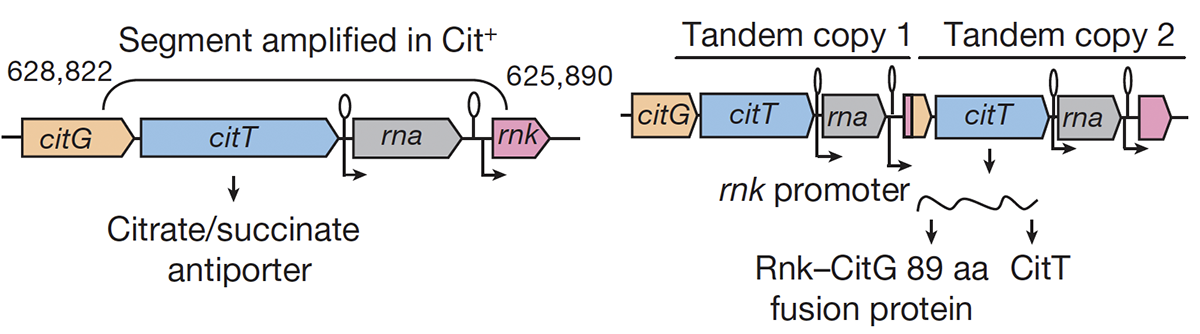
Blount Z D; Barrick J E; Davidson C J; Lenski R E
Genomic analysis of a key innovation in an experimental Escherichia coli population Journal Article
Nature, 489 (7417), pp. 513–518, 2012, ISSN: 0028-0836.
Abstract | Links | BibTeX | Altmetric | Tags: Citrate Evolution, Genome Evolution, Genotypes and Phenotypes, Historical Contingency, Mutation Rates
@article{Blount2012,
title = {Genomic analysis of a key innovation in an experimental \emph{Escherichia coli} population},
author = {Zachary D. Blount and Jeffrey E. Barrick and Carla J. Davidson and Richard E. Lenski},
url = {http://www.nature.com/articles/nature11514},
doi = {10.1038/nature11514},
issn = {0028-0836},
year = {2012},
date = {2012-09-01},
urldate = {2012-09-01},
journal = {Nature},
volume = {489},
number = {7417},
pages = {513--518},
abstract = {Evolutionary novelties have been important in the history of life, but their origins are usually difficult to examine in detail. We previously described the evolution of a novel trait, aerobic citrate utilization (Cit +), in an experimental population of \textit{Escherichia coli}. Here we analyse genome sequences to investigate the history and genetic basis of this trait. At least three distinct clades coexisted for more than 10,000 generations before its emergence. The Cit + trait originated in one clade by a tandem duplication that captured an aerobically expressed promoter for the expression of a previously silent citrate transporter. The clades varied in their propensity to evolve this novel trait, although genotypes able to do so existed in all three clades, implying that multiple potentiating mutations arose during the population's history. Our findings illustrate the importance of promoter capture and altered gene regulation in mediating the exaptation events that often underlie evolutionary innovations.},
keywords = {Citrate Evolution, Genome Evolution, Genotypes and Phenotypes, Historical Contingency, Mutation Rates},
pubstate = {published},
tppubtype = {article}
}